import numpy as np
np.random.seed(20)
import matplotlib.pyplot as plt
import pandas as pd
# Retina display
%config InlineBackend.figure_format = 'retina'
%matplotlib inline
from latexify import latexify, format_axesHyperparams Tuning Strategies Experimentation
ML
Tutorial
MakeMoons Dataset
1.1 Fixed Train-Test (70:30) split ; No Tuning
# Import necessary libraries
from sklearn.model_selection import train_test_split
from sklearn.tree import DecisionTreeClassifier
from sklearn.datasets import make_moons
# Generate the dataset
X, y = make_moons(n_samples=1000, noise=0.3, random_state=42)
# Split the data into training, validation, and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=2)latexify(fig_width=5, fig_height=4)
plt.scatter(X_train[:, 0], X_train[:, 1], c=y_train, label='Train')
plt.scatter(X_test[:, 0], X_test[:, 1], c=y_test, marker='x', label='Test')
format_axes(plt.gca())
plt.legend()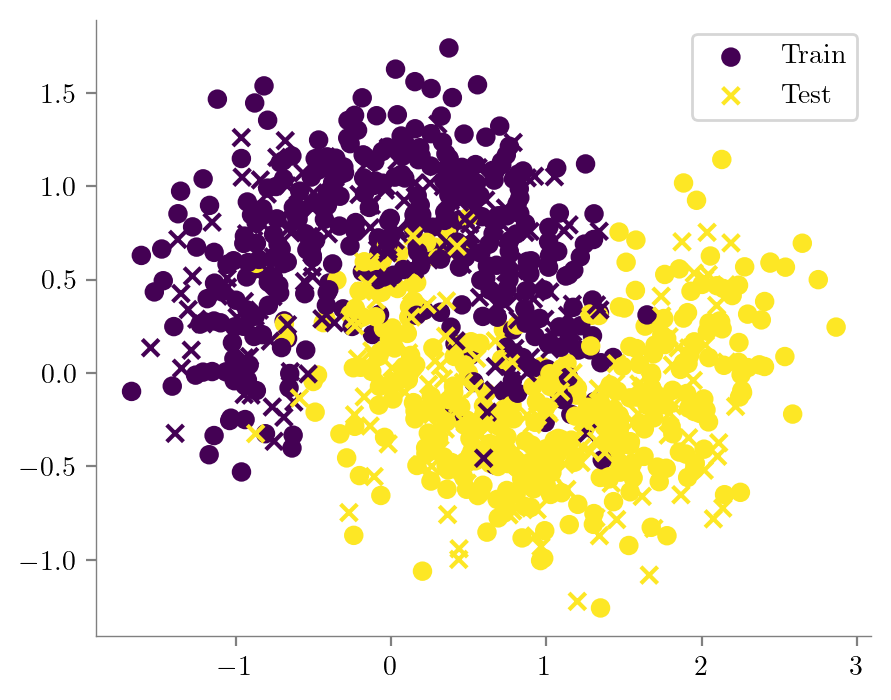
len(X_train), len(X_test)(700, 300)#hyperparameters take their default values
dt_classifier = DecisionTreeClassifier(random_state=42)
dt_classifier.fit(X_train, y_train)
# Make predictions on the test set
test_accuracy = dt_classifier.score(X_test, y_test)
print("Test set accuracy: {:.4f}".format(test_accuracy))Test set accuracy: 0.89331.2 Multiple Random Train-Test splits
# Initialize an empty list to store the accuracy metrics
accuracy_metrics = []
all_test_sets = []
all_predictions = []
X_tests = []
# Perform 10 random train-test splits and repeat the fit
for _ in range(10):
# Split the data into training and test sets
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.3, random_state=np.random.randint(100))
X_tests.append(X_test)
# Create and fit the decision tree classifier
dt_classifier = DecisionTreeClassifier(random_state=42)
dt_classifier.fit(X_train, y_train)
current_predictions = dt_classifier.predict(X_test)
all_predictions.append(current_predictions)
current_accuracy = np.mean(current_predictions == y_test)
all_test_sets.append(y_test)
# Calculate the accuracy on the test set
test_accuracy = dt_classifier.score(X_test, y_test)
# Append the accuracy to the list
accuracy_metrics.append(test_accuracy)
# Calculate the mean and standard deviation of the accuracy metrics
mean_accuracy = np.mean(accuracy_metrics)
std_accuracy = np.std(accuracy_metrics)
# Print the mean and standard deviation
print("Mean accuracy: {:.4f}".format(mean_accuracy))
print("Standard deviation: {:.4f}".format(std_accuracy))
# Print minimum and maximum accuracies
print("Minimum accuracy: {:.4f}".format(min(accuracy_metrics)))
print("Maximum accuracy: {:.4f}".format(max(accuracy_metrics)))Mean accuracy: 0.8800
Standard deviation: 0.0211
Minimum accuracy: 0.8400
Maximum accuracy: 0.9133# Find number of unique element in X_tests
found_unique_test_samples = len(np.unique(np.concatenate(X_tests), axis=0))
print(f"Number of unique test samples: {found_unique_test_samples}")
print(f"Ideally we wanted {len(X)} unique test samples")Number of unique test samples: 964
Ideally we wanted 1000 unique test samples1.3 K-Fold Cross Validation
import numpy as np
# Define the number of folds (k)
k = 5
# Initialize lists to store predictions and accuracies
predictions = {}
accuracies = []
# Calculate the size of each fold
fold_size = len(X) // k
# Perform k-fold cross-validation
for i in range(k):
# Split the data into training and test sets
test_start = i * fold_size
test_end = (i + 1) * fold_size
test_set = X[test_start:test_end]
test_labels = y[test_start:test_end]
training_set = np.concatenate((X[:test_start], X[test_end:]), axis=0)
print(len(test_set))
training_labels = np.concatenate((y[:test_start], y[test_end:]), axis=0)
# Train the model
dt_classifier = DecisionTreeClassifier(random_state=42)
dt_classifier.fit(training_set, training_labels)
# Make predictions on the validation set
fold_predictions = dt_classifier.predict(test_set)
# Calculate the accuracy of the fold
fold_accuracy = np.mean(fold_predictions == test_labels)
# Store the predictions and accuracy of the fold
predictions[i] = fold_predictions
accuracies.append(fold_accuracy)
# Print the predictions and accuracies of each fold
for i in range(k):
print("Fold {}: Accuracy: {:.4f}".format(i+1, accuracies[i]))200
200
200
200
200
Fold 1: Accuracy: 0.8700
Fold 2: Accuracy: 0.8850
Fold 3: Accuracy: 0.9300
Fold 4: Accuracy: 0.8650
Fold 5: Accuracy: 0.8850from cgi import test
from sklearn.model_selection import KFold
# Define the number of folds (k)
k = 5
# Initialize lists to store predictions and accuracies
predictions = {}
accuracies = []
# Create a KFold instance
kf = KFold(n_splits=k, shuffle=False)
# Perform k-fold cross-validation
for i, (train_index, test_index) in enumerate(kf.split(X)):
# Split the data into training and test sets
training_set, test_set = X[train_index], X[test_index]
print(len(test_set))
training_labels, test_labels = y[train_index], y[test_index]
# Train the model
dt_classifier = DecisionTreeClassifier(random_state=42)
dt_classifier.fit(training_set, training_labels)
# Make predictions on the validation set
fold_predictions = dt_classifier.predict(test_set)
# Calculate the accuracy of the fold
fold_accuracy = np.mean(fold_predictions == test_labels)
# Store the predictions and accuracy of the fold
predictions[i] = fold_predictions
accuracies.append(fold_accuracy)
# Print the predictions and accuracy of each fold
print("Fold {}: Accuracy: {:.4f}".format(i+1, fold_accuracy))200
Fold 1: Accuracy: 0.8700
200
Fold 2: Accuracy: 0.8850
200
Fold 3: Accuracy: 0.9300
200
Fold 4: Accuracy: 0.8650
200
Fold 5: Accuracy: 0.8850Micro and Macro Averaging
from sklearn.metrics import accuracy_score
# Method 1 for computing accuracy
accuracy_1 = accuracy_score(y, np.concatenate(list(predictions.values())))
# Calculate macro-averaged accuracy
accuracy_2 = np.mean(accuracies)
# Print the micro and macro averaged accuracy
print("Method 1 accuracy: {:.4f}".format(accuracy_1))
print("Method2 accuracy: {:.4f}".format(accuracy_2))Method 1 accuracy: 0.8870
Method2 accuracy: 0.88702.1 Fixed Train-Test Split (hyperparameters tuned on Validation set)
2.1.1 Validation Set as fixed Subset of Training Set
len(X)1000# Step 1: Split the data into training, validation, and test sets
X_train_val, X_test, y_train_val, y_test = train_test_split(X, y, test_size=0.3, random_state=42)
X_train, X_val, y_train, y_val = train_test_split(X_train_val, y_train_val, test_size=0.285, random_state=42)print("Number of training examples: {}".format(len(X_train)))
print("Number of validation examples: {}".format(len(X_val)))
print("Number of testing examples: {}".format(len(X_test)))Number of training examples: 500
Number of validation examples: 200
Number of testing examples: 300hyperparameters = {}
hyperparameters['max_depth'] = [1,2,3,4,5,6,7,8,9,10]
hyperparameters['min_samples_split'] = [2,3,4,5,6,7,8]
hyperparameters['criteria_values'] = ['gini', 'entropy']
best_accuracy = 0
best_hyperparameters = {}
out = {}
count = 0
for max_depth in hyperparameters['max_depth']:
for min_samples_split in hyperparameters['min_samples_split']:
for criterion in hyperparameters['criteria_values']:
# Create and fit the decision tree classifier with the current hyperparameters
dt_classifier = DecisionTreeClassifier(max_depth=max_depth, min_samples_split=min_samples_split, criterion=criterion, random_state=42)
dt_classifier.fit(X_train, y_train)
# Evaluate the performance on the validation set
val_accuracy = dt_classifier.score(X_val, y_val)
out[count] = {'max_depth': max_depth, 'min_samples_split': min_samples_split, 'criterion': criterion, 'val_accuracy': val_accuracy}
count += 1hparam_df = pd.DataFrame(out).T
hparam_df| max_depth | min_samples_split | criterion | val_accuracy | |
|---|---|---|---|---|
| 0 | 1 | 2 | gini | 0.785 |
| 1 | 1 | 2 | entropy | 0.785 |
| 2 | 1 | 3 | gini | 0.785 |
| 3 | 1 | 3 | entropy | 0.785 |
| 4 | 1 | 4 | gini | 0.785 |
| ... | ... | ... | ... | ... |
| 135 | 10 | 6 | entropy | 0.895 |
| 136 | 10 | 7 | gini | 0.89 |
| 137 | 10 | 7 | entropy | 0.895 |
| 138 | 10 | 8 | gini | 0.885 |
| 139 | 10 | 8 | entropy | 0.895 |
140 rows × 4 columns
hparam_df.sort_values(by='val_accuracy', ascending=False).head(10)| max_depth | min_samples_split | criterion | val_accuracy | |
|---|---|---|---|---|
| 76 | 6 | 5 | gini | 0.925 |
| 77 | 6 | 5 | entropy | 0.925 |
| 78 | 6 | 6 | gini | 0.925 |
| 79 | 6 | 6 | entropy | 0.925 |
| 80 | 6 | 7 | gini | 0.925 |
| 81 | 6 | 7 | entropy | 0.925 |
| 83 | 6 | 8 | entropy | 0.925 |
| 70 | 6 | 2 | gini | 0.92 |
| 82 | 6 | 8 | gini | 0.92 |
| 90 | 7 | 5 | gini | 0.915 |
# Ensure dtype of val_accuracy is float
hparam_df['val_accuracy'] = hparam_df['val_accuracy'].astype(float)best_hyperparameters_row = hparam_df.iloc[hparam_df['val_accuracy'].idxmax()]
best_accuracy = best_hyperparameters_row['val_accuracy']
best_hyperparameters = best_hyperparameters_row[['max_depth', 'min_samples_split', 'criterion']].to_dict()best_hyperparameters{'max_depth': 6, 'min_samples_split': 5, 'criterion': 'gini'}# Evaluate the performance of the selected hyperparameter combination on the test set
dt_classifier = DecisionTreeClassifier(max_depth=best_hyperparameters['max_depth'],
min_samples_split=best_hyperparameters['min_samples_split'],
criterion=best_hyperparameters['criterion'],
random_state=42)
dt_classifier.fit(X_train_val, y_train_val)
test_accuracy = dt_classifier.score(X_test, y_test)
print("Best Hyperparameters:", best_hyperparameters)
print("Validation Set accuracy: {:.4f}".format(best_accuracy))
print("Test Set accuracy: {:.4f}".format(test_accuracy))Best Hyperparameters: {'max_depth': 6, 'min_samples_split': 5, 'criterion': 'gini'}
Validation Set accuracy: 0.9250
Test Set accuracy: 0.9067Avoiding nested loops by using itertools.product
for max_depth in hyperparameters['max_depth']:
for min_samples_split in hyperparameters['min_samples_split']:
for criterion in hyperparameters['criteria_values']:
# Create and fit the decision tree classifier with the current hyperparameters
dt_classifier = DecisionTreeClassifier(max_depth=max_depth, min_samples_split=min_samples_split, criterion=criterion, random_state=42)
dt_classifier.fit(X_train, y_train)
# Evaluate the performance on the validation set
val_accuracy = dt_classifier.score(X_val, y_val)
out[count] = {'max_depth': max_depth, 'min_samples_split': min_samples_split, 'criterion': criterion, 'val_accuracy': val_accuracy}
count += 1from itertools import product
for max_depth, min_samples_split, criterion in product(hyperparameters['max_depth'], hyperparameters['min_samples_split'], hyperparameters['criteria_values']):
# Define the Decision Tree Classifier
dt_classifier = DecisionTreeClassifier(
max_depth=max_depth,
min_samples_split=min_samples_split,
criterion=criterion,
random_state=42
)
dt_classifier.fit(X_train, y_train)2.1.2 Multiple random subsets of Training Set used as Validation Set
# Initialize a list to store the optimal hyperparameters for each validation set
optimal_hyperparameters = {}
test_accuracies = []
# Set the number of subsets and iterations
num_subsets = 5
# Make a pandas dataframe with columns as the hyperparameters, subset number, and validation accuracy
hyperparameters_df = pd.DataFrame(columns=['max_depth', 'min_samples_split', 'criterion', 'subset', 'validation accuracy'])
# Iterate over the subsets
for i in range(num_subsets):
# Split the data into training and validation sets
X_train_subset, X_val_subset, y_train_subset, y_val_subset = train_test_split(X_train_val, y_train_val, test_size=0.285, random_state=i)
# Initialize variables to store the best hyperparameters and accuracy for the current subset
best_accuracy = 0
best_hyperparameters = {}
# Iterate over the hyperparameter values
for max_depth in hyperparameters['max_depth']:
for min_samples_split in hyperparameters['min_samples_split']:
for criterion in hyperparameters['criteria_values']:
# Initialize and train the model with the current hyperparameters
dt_classifier = DecisionTreeClassifier(max_depth=max_depth, min_samples_split=min_samples_split, criterion=criterion, random_state=42)
dt_classifier.fit(X_train_subset, y_train_subset)
# Evaluate the model on the validation set
val_accuracy = dt_classifier.score(X_val_subset, y_val_subset)
hyperparameters_df.loc[len(hyperparameters_df)] = [max_depth, min_samples_split, criterion, i+1, val_accuracy]
# Update the best accuracy and hyperparameters
if val_accuracy > best_accuracy:
best_accuracy = val_accuracy
best_hyperparameters = {
'max_depth': max_depth,
'min_samples_split': min_samples_split,
'criterion': criterion
}
optimal_hyperparameters[i] = best_hyperparameters
# Evaluate the model with the best hyperparameters on the test set
dt_classifier = DecisionTreeClassifier(max_depth=best_hyperparameters['max_depth'], min_samples_split=best_hyperparameters['min_samples_split'], criterion=best_hyperparameters['criterion'], random_state=42)
dt_classifier.fit(X_train_val, y_train_val)
test_accuracy = dt_classifier.score(X_test, y_test)
test_accuracies.append(test_accuracy)
print("Optimal hyperparameters for {} inner folds/validation sets".format(num_subsets))
print()
# Print the optimal hyperparameters for each validation set
for i in range(num_subsets):
print("Optimal hyperparameters for validation set {}: {}".format(i+1, optimal_hyperparameters[i]))
print("Test Accuracy for validation set {}: {:.4f}".format(i+1, test_accuracies[i]))Optimal hyperparameters for 5 inner folds/validation sets
Optimal hyperparameters for validation set 1: {'max_depth': 7, 'min_samples_split': 6, 'criterion': 'entropy'}
Test Accuracy for validation set 1: 0.9000
Optimal hyperparameters for validation set 2: {'max_depth': 5, 'min_samples_split': 7, 'criterion': 'gini'}
Test Accuracy for validation set 2: 0.9033
Optimal hyperparameters for validation set 3: {'max_depth': 6, 'min_samples_split': 2, 'criterion': 'entropy'}
Test Accuracy for validation set 3: 0.9233
Optimal hyperparameters for validation set 4: {'max_depth': 7, 'min_samples_split': 4, 'criterion': 'entropy'}
Test Accuracy for validation set 4: 0.9000
Optimal hyperparameters for validation set 5: {'max_depth': 6, 'min_samples_split': 2, 'criterion': 'entropy'}
Test Accuracy for validation set 5: 0.9233hyperparameters_df| max_depth | min_samples_split | criterion | subset | validation accuracy | |
|---|---|---|---|---|---|
| 0 | 1 | 2 | gini | 1 | 0.790 |
| 1 | 1 | 2 | entropy | 1 | 0.790 |
| 2 | 1 | 3 | gini | 1 | 0.790 |
| 3 | 1 | 3 | entropy | 1 | 0.790 |
| 4 | 1 | 4 | gini | 1 | 0.790 |
| ... | ... | ... | ... | ... | ... |
| 695 | 10 | 6 | entropy | 5 | 0.900 |
| 696 | 10 | 7 | gini | 5 | 0.905 |
| 697 | 10 | 7 | entropy | 5 | 0.900 |
| 698 | 10 | 8 | gini | 5 | 0.905 |
| 699 | 10 | 8 | entropy | 5 | 0.900 |
700 rows × 5 columns
grouped_df = hyperparameters_df.groupby(['max_depth', 'min_samples_split', 'criterion']).mean()['validation accuracy']
grouped_dfmax_depth min_samples_split criterion
1 2 entropy 0.769
gini 0.771
3 entropy 0.769
gini 0.771
4 entropy 0.769
...
10 6 gini 0.889
7 entropy 0.902
gini 0.894
8 entropy 0.904
gini 0.893
Name: validation accuracy, Length: 140, dtype: float64grouped_df.sort_values(ascending=False).head(10)max_depth min_samples_split criterion
6 7 entropy 0.914
8 entropy 0.914
7 7 entropy 0.912
6 6 entropy 0.912
7 8 entropy 0.912
6 entropy 0.910
6 4 entropy 0.910
5 entropy 0.910
7 4 entropy 0.909
5 entropy 0.909
Name: validation accuracy, dtype: float64optimal_hyperparams = grouped_df.idxmax()
optimal_hyperparams(6, 7, 'entropy')df_classifier = DecisionTreeClassifier(max_depth=optimal_hyperparams[0], min_samples_split=optimal_hyperparams[1], criterion=optimal_hyperparams[2], random_state=42)
df_classifier.fit(X_train_val, y_train_val)
test_accuracy = df_classifier.score(X_test, y_test)
print("Test accuracy: {:.4f}".format(test_accuracy))Test accuracy: 0.92332.2 Nested Cross-Validation
hyperparameters['max_depth'] = [1,2,3,4,5,6,7,8,9,10]
hyperparameters['min_samples_split'] = [2,3,4,5,6,7,8]
hyperparameters['criteria_values'] = ['gini', 'entropy']num_outer_folds = 5
num_inner_folds = 5
kf_outer = KFold(n_splits=num_outer_folds, shuffle=False)
kf_inner = KFold(n_splits=num_inner_folds, shuffle=False)
# Initialize lists to store the accuracies for the outer and inner loops
outer_loop_accuracies = []
inner_loop_accuracies = []
results= {}
outer_count = 0
overall_count = 0
# Iterate over the outer folds
for outer_train_index, outer_test_index in kf_outer.split(X):
# Split the data into outer training and test sets
X_outer_train, X_outer_test = X[outer_train_index], X[outer_test_index]
y_outer_train, y_outer_test = y[outer_train_index], y[outer_test_index]
inner_count = 0
for innner_train_index, inner_test_index in kf_inner.split(X_outer_train):
print("*****"*20)
print("Outer Fold {}, Inner Fold {}".format(outer_count+1, inner_count+1))
# Split the data into inner training and test sets
X_inner_train, X_inner_test = X_outer_train[innner_train_index], X_outer_train[inner_test_index]
y_inner_train, y_inner_test = y_outer_train[innner_train_index], y_outer_train[inner_test_index]
for max_depth, min_samples_split, criterion in product(hyperparameters['max_depth'],
hyperparameters['min_samples_split'],
hyperparameters['criteria_values']):
#print(max_depth, min_samples_split, criterion)
# Initialize and train the model with the current hyperparameters
dt_classifier = DecisionTreeClassifier(max_depth=max_depth,
min_samples_split=min_samples_split,
criterion=criterion, random_state=42)
dt_classifier.fit(X_inner_train, y_inner_train)
# Evaluate the model on the inner test set
val_accuracy = dt_classifier.score(X_inner_test, y_inner_test)
results[overall_count] = {'outer_fold': outer_count,
'inner_fold': inner_count,
'max_depth': max_depth,
'min_samples_split': min_samples_split,
'criterion': criterion,
'val_accuracy': val_accuracy}
overall_count += 1
inner_count += 1
outer_count += 1
****************************************************************************************************
Outer Fold 1, Inner Fold 1
****************************************************************************************************
Outer Fold 1, Inner Fold 2
****************************************************************************************************
Outer Fold 1, Inner Fold 3
****************************************************************************************************
Outer Fold 1, Inner Fold 4
****************************************************************************************************
Outer Fold 1, Inner Fold 5
****************************************************************************************************
Outer Fold 2, Inner Fold 1
****************************************************************************************************
Outer Fold 2, Inner Fold 2
****************************************************************************************************
Outer Fold 2, Inner Fold 3
****************************************************************************************************
Outer Fold 2, Inner Fold 4
****************************************************************************************************
Outer Fold 2, Inner Fold 5
****************************************************************************************************
Outer Fold 3, Inner Fold 1
****************************************************************************************************
Outer Fold 3, Inner Fold 2
****************************************************************************************************
Outer Fold 3, Inner Fold 3
****************************************************************************************************
Outer Fold 3, Inner Fold 4
****************************************************************************************************
Outer Fold 3, Inner Fold 5
****************************************************************************************************
Outer Fold 4, Inner Fold 1
****************************************************************************************************
Outer Fold 4, Inner Fold 2
****************************************************************************************************
Outer Fold 4, Inner Fold 3
****************************************************************************************************
Outer Fold 4, Inner Fold 4
****************************************************************************************************
Outer Fold 4, Inner Fold 5
****************************************************************************************************
Outer Fold 5, Inner Fold 1
****************************************************************************************************
Outer Fold 5, Inner Fold 2
****************************************************************************************************
Outer Fold 5, Inner Fold 3
****************************************************************************************************
Outer Fold 5, Inner Fold 4
****************************************************************************************************
Outer Fold 5, Inner Fold 5overall_results = pd.DataFrame(results).Toverall_results| outer_fold | inner_fold | max_depth | min_samples_split | criterion | val_accuracy | |
|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 1 | 2 | gini | 0.7625 |
| 1 | 0 | 0 | 1 | 2 | entropy | 0.7625 |
| 2 | 0 | 0 | 1 | 3 | gini | 0.7625 |
| 3 | 0 | 0 | 1 | 3 | entropy | 0.7625 |
| 4 | 0 | 0 | 1 | 4 | gini | 0.7625 |
| ... | ... | ... | ... | ... | ... | ... |
| 3495 | 4 | 4 | 10 | 6 | entropy | 0.9 |
| 3496 | 4 | 4 | 10 | 7 | gini | 0.91875 |
| 3497 | 4 | 4 | 10 | 7 | entropy | 0.9 |
| 3498 | 4 | 4 | 10 | 8 | gini | 0.925 |
| 3499 | 4 | 4 | 10 | 8 | entropy | 0.9 |
3500 rows × 6 columns
Find the best hyperparameters for each outer fold
outer_fold = 0
outer_fold_df = overall_results.query('outer_fold == @outer_fold')
outer_fold_df| outer_fold | inner_fold | max_depth | min_samples_split | criterion | val_accuracy | |
|---|---|---|---|---|---|---|
| 0 | 0 | 0 | 1 | 2 | gini | 0.7625 |
| 1 | 0 | 0 | 1 | 2 | entropy | 0.7625 |
| 2 | 0 | 0 | 1 | 3 | gini | 0.7625 |
| 3 | 0 | 0 | 1 | 3 | entropy | 0.7625 |
| 4 | 0 | 0 | 1 | 4 | gini | 0.7625 |
| ... | ... | ... | ... | ... | ... | ... |
| 695 | 0 | 4 | 10 | 6 | entropy | 0.85 |
| 696 | 0 | 4 | 10 | 7 | gini | 0.86875 |
| 697 | 0 | 4 | 10 | 7 | entropy | 0.85625 |
| 698 | 0 | 4 | 10 | 8 | gini | 0.86875 |
| 699 | 0 | 4 | 10 | 8 | entropy | 0.85625 |
700 rows × 6 columns
Aggregate the validation accuracies for each hyperparameter combination across all inner folds
outer_fold_df.groupby(['max_depth', 'min_samples_split', 'criterion']).mean()['val_accuracy'].sort_values(ascending=False).head(10)max_depth min_samples_split criterion
6 7 gini 0.9175
8 gini 0.9175
6 gini 0.9175
4 gini 0.91625
3 gini 0.91625
2 gini 0.91625
5 gini 0.91625
7 6 gini 0.91625
7 gini 0.91625
8 gini 0.915
Name: val_accuracy, dtype: object