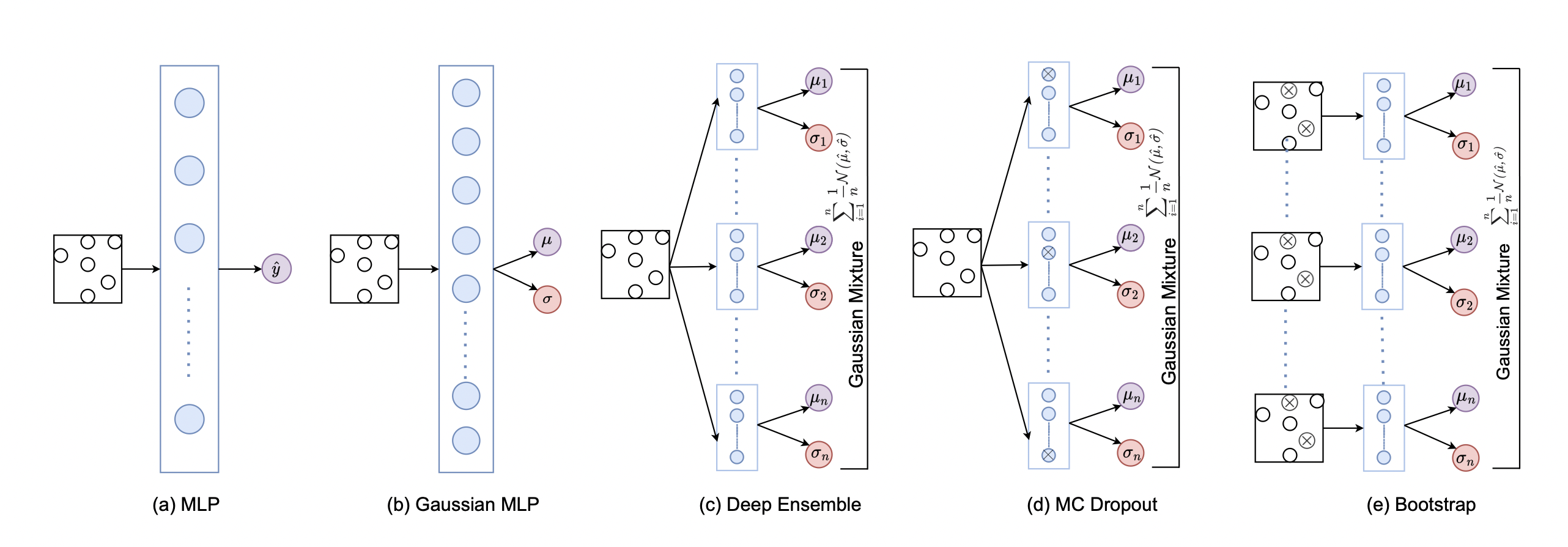
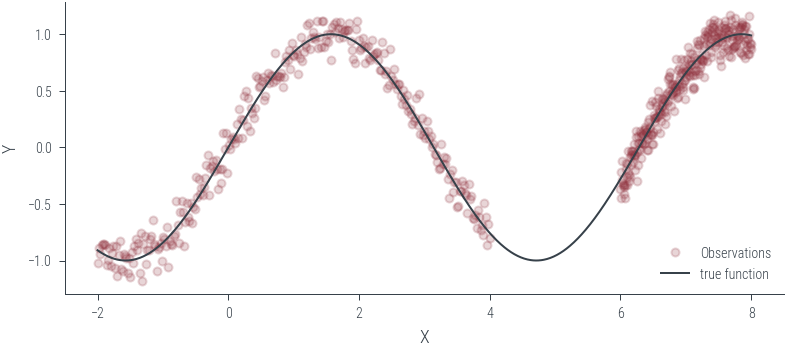
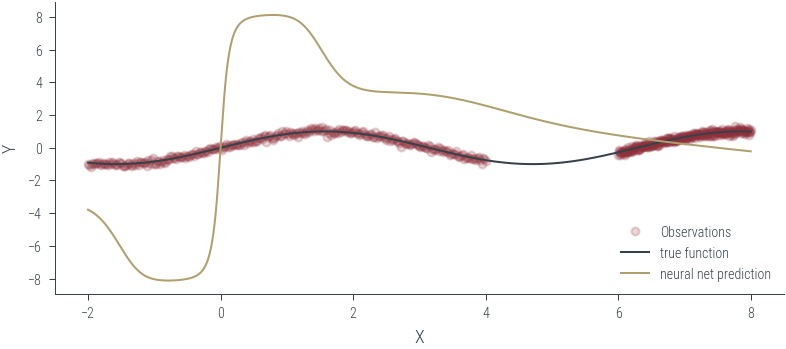
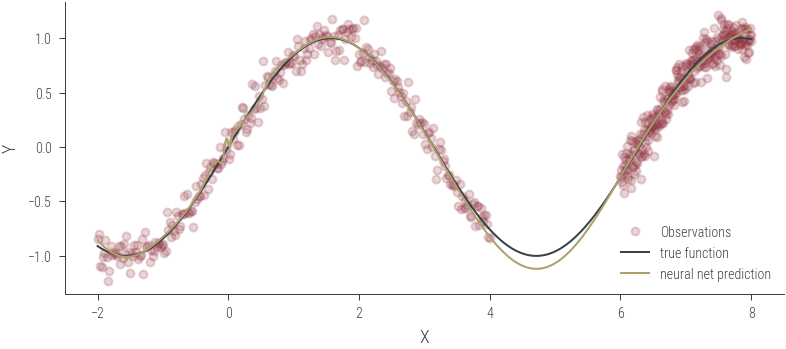
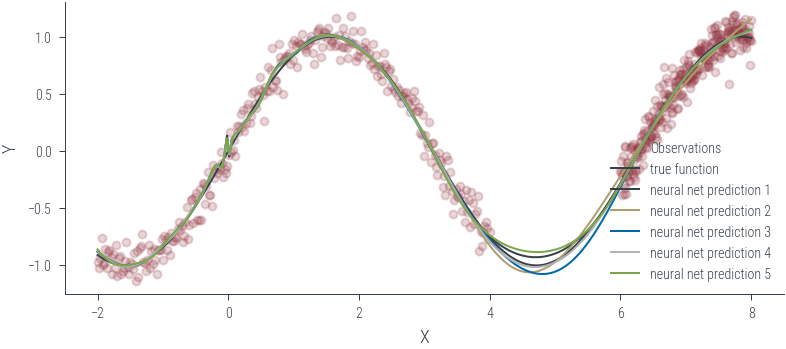
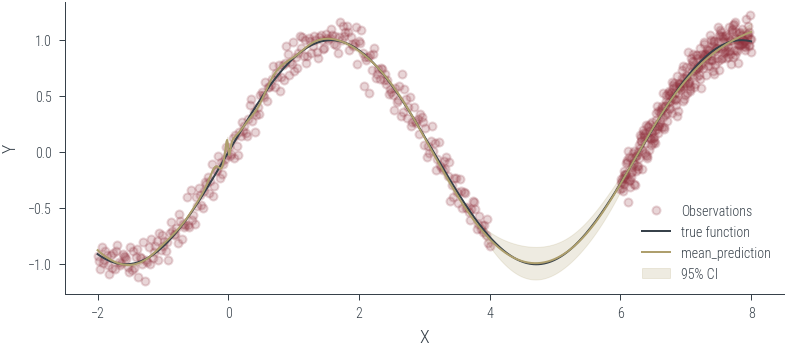
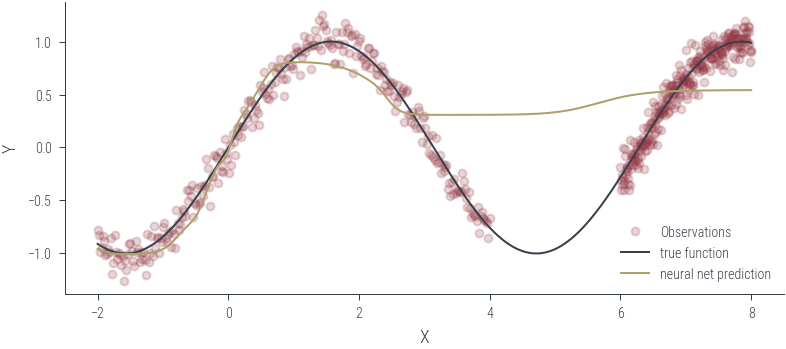
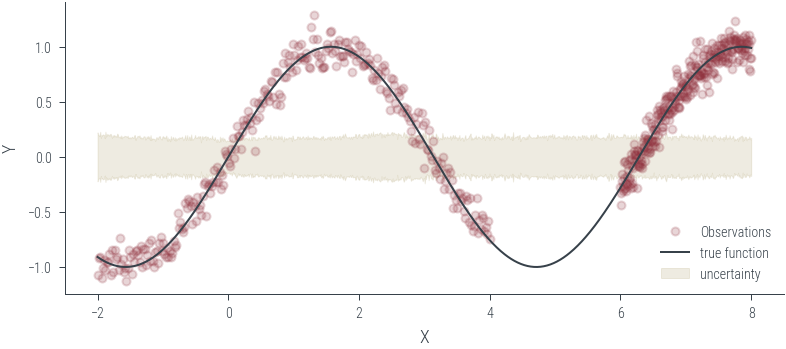
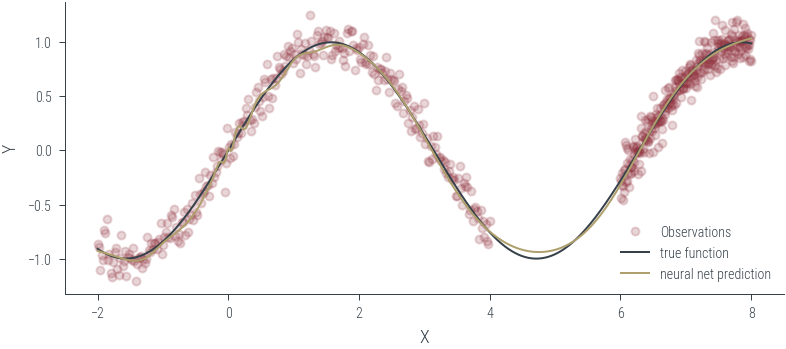
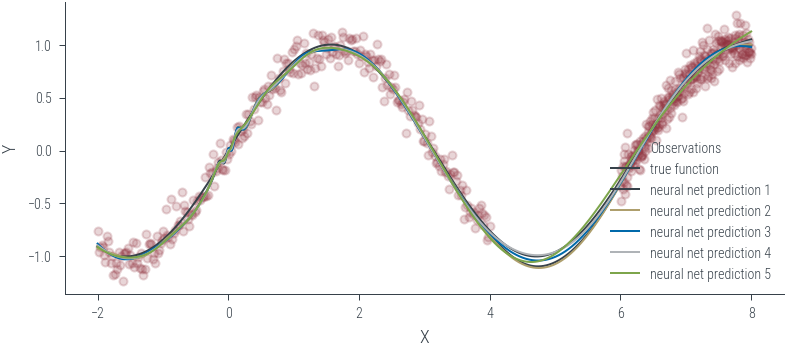
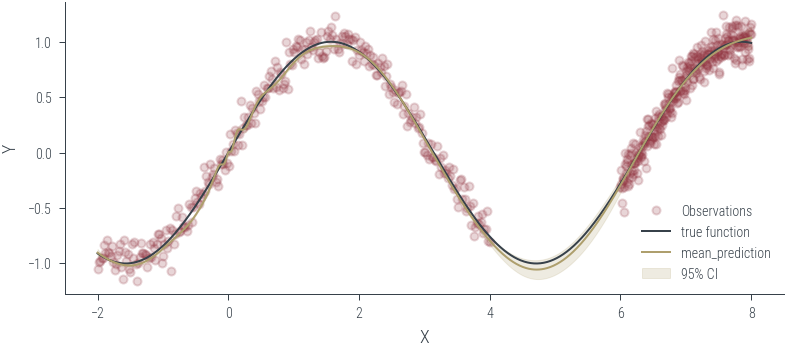
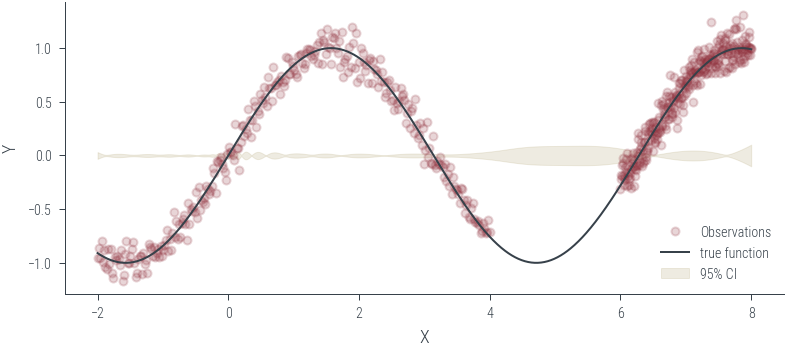
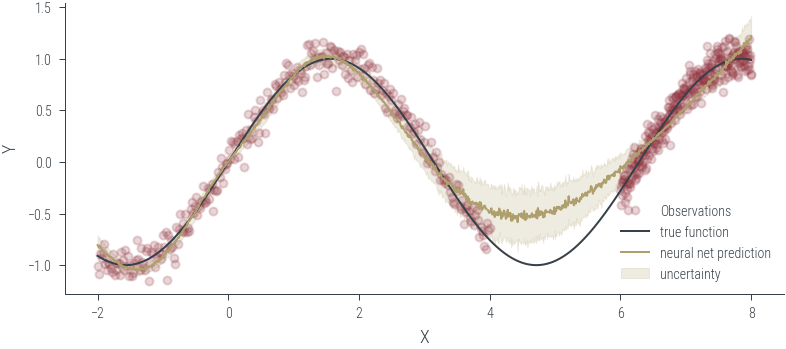
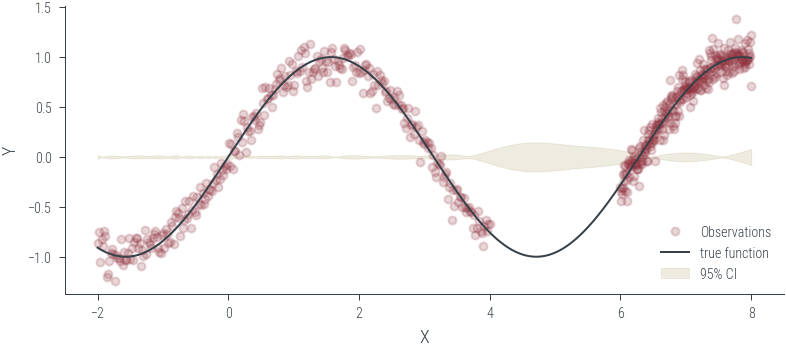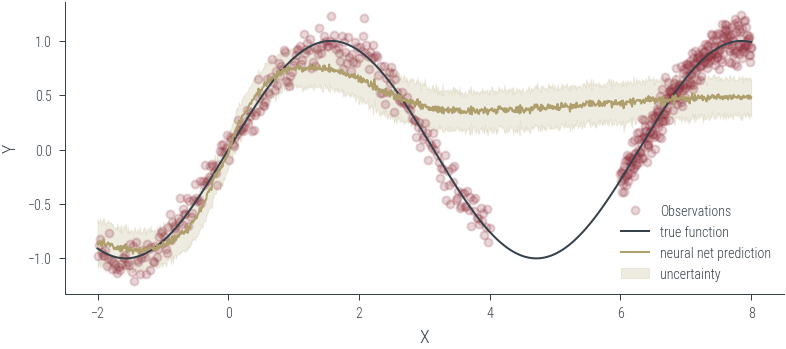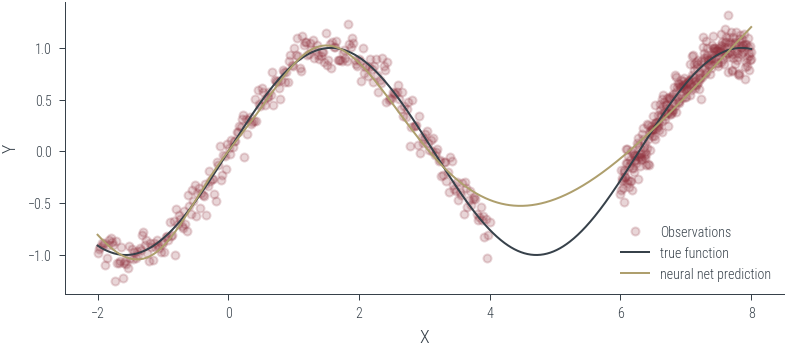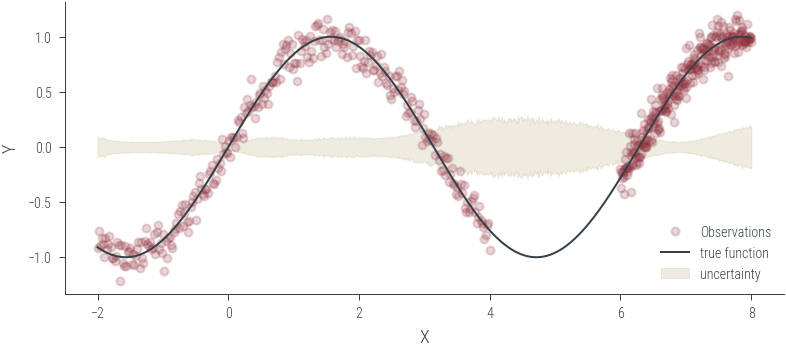Training ensemble member 0
Epoch 0, loss 3.468
Epoch 300, loss 0.038
Epoch 600, loss 0.020
Epoch 900, loss 0.014
Epoch 1200, loss 0.012
Epoch 1500, loss 0.011
Epoch 1800, loss 0.011
Epoch 2100, loss 0.010
Epoch 2400, loss 0.010
Epoch 2700, loss 0.010
Epoch 3000, loss 0.010
Epoch 3300, loss 0.010
Epoch 3600, loss 0.010
Epoch 3900, loss 0.010
Epoch 4200, loss 0.010
Epoch 4500, loss 0.010
Epoch 4800, loss 0.010
Epoch 5100, loss 0.010
Epoch 5400, loss 0.009
Epoch 5700, loss 0.009
Training ensemble member 1
Epoch 0, loss 34.524
Epoch 300, loss 0.042
Epoch 600, loss 0.022
Epoch 900, loss 0.015
Epoch 1200, loss 0.014
Epoch 1500, loss 0.013
Epoch 1800, loss 0.013
Epoch 2100, loss 0.013
Epoch 2400, loss 0.013
Epoch 2700, loss 0.012
Epoch 3000, loss 0.012
Epoch 3300, loss 0.012
Epoch 3600, loss 0.012
Epoch 3900, loss 0.012
Epoch 4200, loss 0.012
Epoch 4500, loss 0.012
Epoch 4800, loss 0.012
Epoch 5100, loss 0.011
Epoch 5400, loss 0.011
Epoch 5700, loss 0.011
Training ensemble member 2
Epoch 0, loss 8.979
Epoch 300, loss 0.019
Epoch 600, loss 0.013
Epoch 900, loss 0.011
Epoch 1200, loss 0.011
Epoch 1500, loss 0.010
Epoch 1800, loss 0.010
Epoch 2100, loss 0.010
Epoch 2400, loss 0.010
Epoch 2700, loss 0.010
Epoch 3000, loss 0.010
Epoch 3300, loss 0.010
Epoch 3600, loss 0.010
Epoch 3900, loss 0.010
Epoch 4200, loss 0.010
Epoch 4500, loss 0.010
Epoch 4800, loss 0.010
Epoch 5100, loss 0.010
Epoch 5400, loss 0.010
Epoch 5700, loss 0.010
Training ensemble member 3
Epoch 0, loss 51.558
Epoch 300, loss 0.151
Epoch 600, loss 0.070
Epoch 900, loss 0.019
Epoch 1200, loss 0.013
Epoch 1500, loss 0.012
Epoch 1800, loss 0.011
Epoch 2100, loss 0.011
Epoch 2400, loss 0.010
Epoch 2700, loss 0.010
Epoch 3000, loss 0.010
Epoch 3300, loss 0.010
Epoch 3600, loss 0.010
Epoch 3900, loss 0.010
Epoch 4200, loss 0.010
Epoch 4500, loss 0.010
Epoch 4800, loss 0.010
Epoch 5100, loss 0.009
Epoch 5400, loss 0.009
Epoch 5700, loss 0.009
Training ensemble member 4
Epoch 0, loss 12.525
Epoch 300, loss 0.100
Epoch 600, loss 0.062
Epoch 900, loss 0.021
Epoch 1200, loss 0.015
Epoch 1500, loss 0.013
Epoch 1800, loss 0.012
Epoch 2100, loss 0.011
Epoch 2400, loss 0.010
Epoch 2700, loss 0.010
Epoch 3000, loss 0.010
Epoch 3300, loss 0.010
Epoch 3600, loss 0.010
Epoch 3900, loss 0.010
Epoch 4200, loss 0.010
Epoch 4500, loss 0.010
Epoch 4800, loss 0.010
Epoch 5100, loss 0.010
Epoch 5400, loss 0.010
Epoch 5700, loss 0.010