import torch
import numpy as np
import matplotlib.pyplot as plt
import pandas as pd
import jax.tree_util as jtu
%matplotlib inline
# Retina display
%config InlineBackend.figure_format = 'retina'
# PyTorch device CUDA0
device = torch.device("cuda:0" if torch.cuda.is_available() else "cpu")Capturing uncertainty in neural nets:
- aleatoric
- homoskedastic (fixed)
- homoskedastic (learnt)
- heteroskedastic
- epistemic uncertainty (Laplace approximation)
- both aleatoric and epistemic uncertainty
Imports
from tueplots import bundles
plt.rcParams.update(bundles.beamer_moml())
# Also add despine to the bundle using rcParams
plt.rcParams["axes.spines.right"] = False
plt.rcParams["axes.spines.top"] = False
# Increase font size to match Beamer template
plt.rcParams["font.size"] = 16
# Make background transparent
plt.rcParams["figure.facecolor"] = "none"Notation

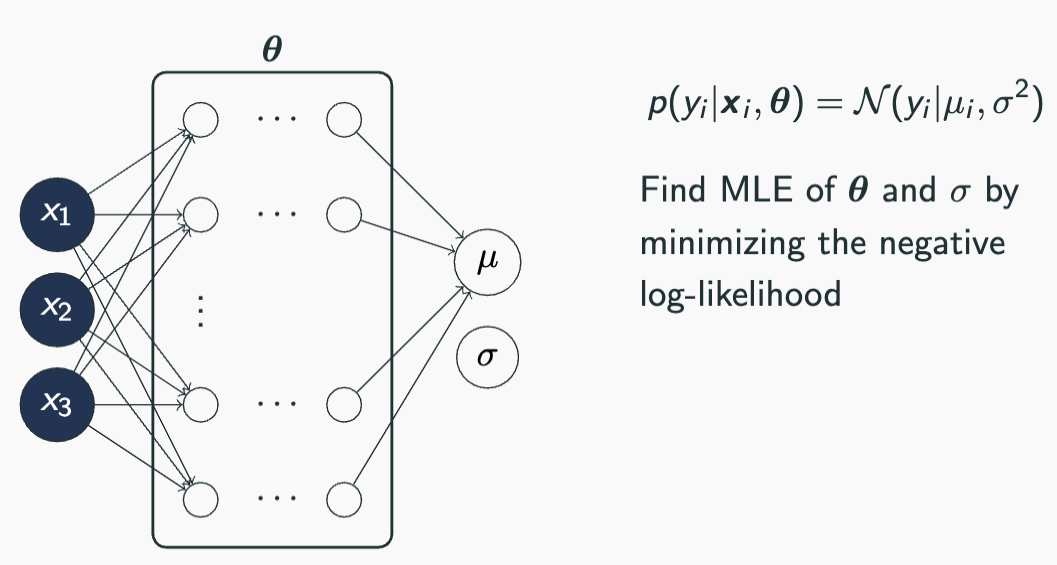
Models capturing aleatoric uncertainty
Dataset containing homoskedastic noise
# Set a fixed random seed for reproducibility
torch.manual_seed(42)
# Define the number of data points
N = 100
# Create a linearly spaced range of values from -1 to 1
x_lin = torch.linspace(-1, 1, N)
# Define a function 'f' to model the data
f = lambda x: 0.5 * x**2 + 0.25 * x**3
# Generate random noise 'eps' with a standard deviation of 0.2
eps = torch.randn(N) * 0.2
# Add noise to the true function to simulate real-world data
y = f(x_lin) + eps
# Move data to the GPU if available (assuming 'device' is defined)
x_lin = x_lin.to(device)
y = y.to(device)# Plot data and true function
plt.plot(x_lin.cpu(), y.cpu(), "o", label="data")
plt.plot(x_lin.cpu(), f(x_lin).cpu(), label="true function")
plt.xlabel("x")
plt.ylabel("y")
plt.legend()<matplotlib.legend.Legend at 0x7f613c0934f0>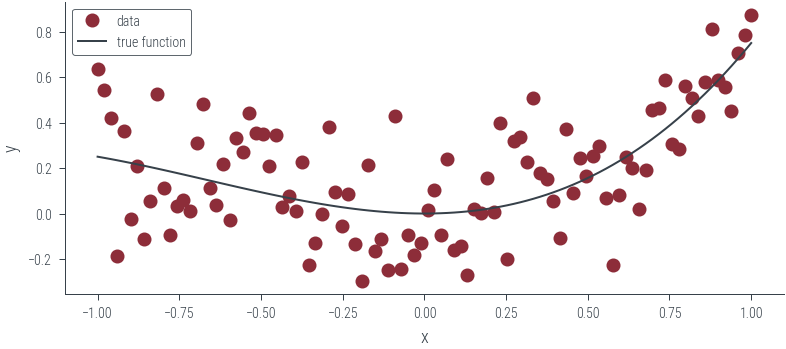
Case 1.1: Models assuming Homoskedastic noise
Case 1.1.1: Homoskedastic noise is fixed beforehand and not learned
class MeanEstimateNN(torch.nn.Module):
def __init__(self, n_hidden=4):
super().__init__()
self.fc1 = torch.nn.Linear(1, n_hidden)
self.fc2 = torch.nn.Linear(n_hidden, n_hidden)
self.fc3 = torch.nn.Linear(n_hidden, 1)
def forward(self, x):
x = self.fc1(x)
x = torch.relu(x)
x = self.fc2(x)
x = torch.relu(x)
mu_hat = self.fc3(x)
return mu_hatdef loss_homoskedastic_noise(model, x, y, params):
"""
Compute the Negative Log Likelihood (NLL) loss for data with Homoskedastic Noise.
It assumes that the noise standard deviation is provided as a parameter.
Args:
model (torch.nn.Module): The neural network model used for mean estimation.
x (torch.Tensor): Input data tensor.
y (torch.Tensor): Target data tensor.
params (dict): A dictionary containing model parameters, including 'log_noise_std'.
Returns:
torch.Tensor: The negative log likelihood loss for the given data.
"""
# Extract the log noise standard deviation from the parameters
log_noise_std = params["log_noise_std"]
# Compute the estimated mean using the model
mu_hat = model(x).squeeze()
# Ensure the shapes of mu_hat and y are compatible
assert mu_hat.shape == y.shape
# Compute the noise standard deviation based on the log value
noise_std = torch.exp(log_noise_std).expand_as(mu_hat)
# Create a Normal distribution with mean mu_hat and standard deviation noise_std
dist = torch.distributions.Normal(mu_hat, noise_std)
# Calculate the negative log likelihood loss and take the mean
return -dist.log_prob(y).mean()homoskedastic_model_fixed_noise = MeanEstimateNN().to(device)
homoskedastic_model_fixed_noiseMeanEstimateNN(
(fc1): Linear(in_features=1, out_features=4, bias=True)
(fc2): Linear(in_features=4, out_features=4, bias=True)
(fc3): Linear(in_features=4, out_features=1, bias=True)
)fixed_log_noise_std = torch.log(torch.tensor(0.5)).to(device)
params = {
"nn_params": homoskedastic_model_fixed_noise.state_dict(),
"log_noise_std": fixed_log_noise_std,
}
loss_homoskedastic_noise(homoskedastic_model_fixed_noise, x_lin[:, None], y, params)tensor(0.7089, device='cuda:0', grad_fn=<NegBackward0>)def plot_results(y_hat, epistemic_std=None, aleatoric_std=None, model_name=""):
plt.scatter(x_lin.cpu(), y.cpu(), s=10, color="C0", label="Data")
plt.plot(x_lin.cpu(), f(x_lin.cpu()), color="C1", label="True function")
plt.plot(x_lin.cpu(), y_hat.cpu(), color="C2", label=model_name)
if epistemic_std is not None:
plt.fill_between(
x_lin.cpu(),
(y_hat - 2 * epistemic_std).cpu(),
(y_hat + 2 * epistemic_std).cpu(),
alpha=0.3,
color="C3",
label="Epistemic uncertainty",
)
if aleatoric_std is not None:
plt.fill_between(
x_lin.cpu(),
(y_hat - 2 * aleatoric_std).cpu(),
(y_hat + 2 * aleatoric_std).cpu(),
alpha=0.3,
color="C2",
label="Aleatoric uncertainty",
)
plt.xlabel("$x$")
plt.ylabel("$y$")
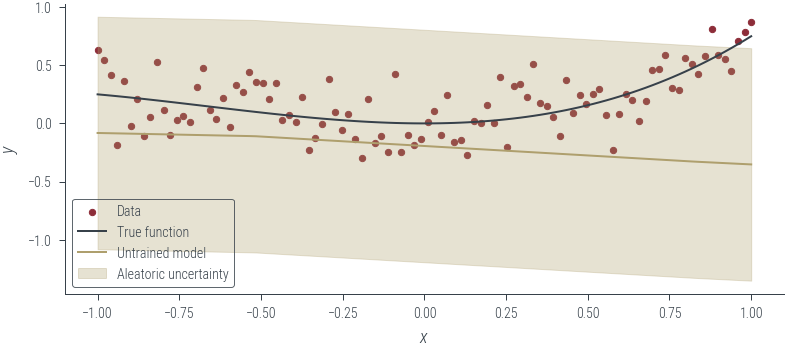
plt.legend()with torch.no_grad():
y_hat = homoskedastic_model_fixed_noise(x_lin[:, None]).squeeze()
plot_results(
y_hat, aleatoric_std=torch.exp(fixed_log_noise_std), model_name="Untrained model"
)
def train_fn(model, loss_func, params, x, y, n_epochs=1000, lr=0.01):
parameter_leaves = jtu.tree_leaves(params)
optimizer = torch.optim.Adam(parameter_leaves, lr=lr)
for epoch in range(n_epochs):
optimizer.zero_grad()
loss = loss_func(model, x, y, params)
loss.backward()
optimizer.step()
# Print every 10 epochs
if epoch % 50 == 0:
print(f"Epoch {epoch}: loss {loss.item():.3f}")
return loss.item()homoskedastic_model_fixed_noise = MeanEstimateNN().to(device)
params = {
"nn_params": list(homoskedastic_model_fixed_noise.parameters()),
"log_noise_std": fixed_log_noise_std,
}train_fn(
homoskedastic_model_fixed_noise,
loss_homoskedastic_noise,
params,
x_lin[:, None],
y,
n_epochs=1000,
lr=0.001,
)Epoch 0: loss 0.369
Epoch 50: loss 0.367
Epoch 100: loss 0.362
Epoch 150: loss 0.352
Epoch 200: loss 0.338
Epoch 250: loss 0.324
Epoch 300: loss 0.312
Epoch 350: loss 0.307
Epoch 400: loss 0.305
Epoch 450: loss 0.304
Epoch 500: loss 0.304
Epoch 550: loss 0.303
Epoch 600: loss 0.303
Epoch 650: loss 0.303
Epoch 700: loss 0.303
Epoch 750: loss 0.303
Epoch 800: loss 0.302
Epoch 850: loss 0.302
Epoch 900: loss 0.302
Epoch 950: loss 0.3020.3021060824394226with torch.no_grad():
y_hat = homoskedastic_model_fixed_noise(x_lin[:, None]).squeeze()
aleatoric_std = torch.exp(fixed_log_noise_std)
plot_results(y_hat, aleatoric_std=aleatoric_std, model_name="Trained model")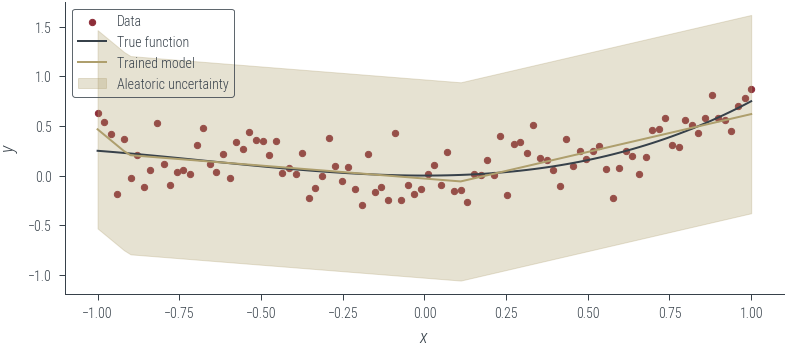
Case 1.1.2: Homoskedastic noise is learnt from the data
The model is the same as in case 1.1.1, but the noise is learned from the data.
homoskedastic_model_learnable_noise = MeanEstimateNN().to(device)
# We keep the noise_std as a parameter, instead of a fixed value
# Earlier code was: fixed_log_noise_std = torch.log(torch.tensor(0.5)).to(device)
log_noise_std = torch.nn.Parameter(torch.tensor(0.0).to(device))
homoskedastic_model_learnable_noiseMeanEstimateNN(
(fc1): Linear(in_features=1, out_features=4, bias=True)
(fc2): Linear(in_features=4, out_features=4, bias=True)
(fc3): Linear(in_features=4, out_features=1, bias=True)
)# Plot the untrained model
with torch.no_grad():
y_hat = homoskedastic_model_learnable_noise(x_lin[:, None]).squeeze()
aleatoric_std = torch.exp(log_noise_std)
plot_results(y_hat, aleatoric_std=aleatoric_std, model_name="Untrained model")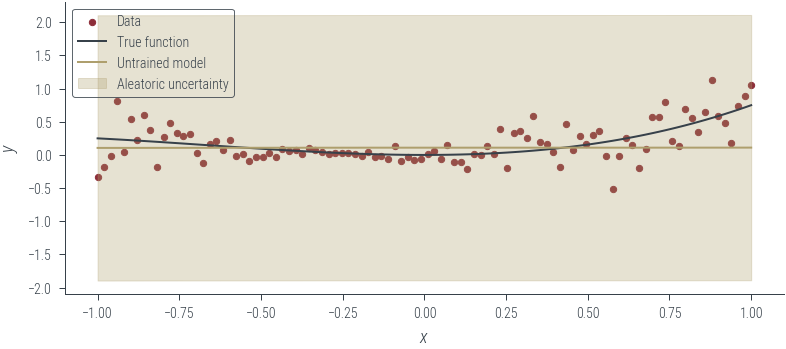
# Train the model
homoskedastic_model_learnable_noise = MeanEstimateNN().to(device)
params = {
"nn_params": list(homoskedastic_model_learnable_noise.parameters()),
"log_noise_std": log_noise_std,
}jtu.tree_leaves(params) [Parameter containing:
tensor(0., device='cuda:0', requires_grad=True),
Parameter containing:
tensor([[-0.7584],
[-0.9339],
[ 0.0176],
[ 0.9118]], device='cuda:0', requires_grad=True),
Parameter containing:
tensor([ 0.5769, -0.5822, -0.1298, -0.7372], device='cuda:0',
requires_grad=True),
Parameter containing:
tensor([[-0.2412, 0.0905, 0.2723, 0.4142],
[-0.4591, 0.3343, -0.3526, 0.1872],
[ 0.4231, 0.0070, 0.4549, -0.4260],
[-0.1910, 0.2916, -0.1089, -0.1024]], device='cuda:0',
requires_grad=True),
Parameter containing:
tensor([-0.2084, 0.3447, 0.2453, 0.1602], device='cuda:0',
requires_grad=True),
Parameter containing:
tensor([[-0.2810, -0.4059, 0.0541, 0.1481]], device='cuda:0',
requires_grad=True),
Parameter containing:
tensor([-0.2309], device='cuda:0', requires_grad=True)][log_noise_std] + list(homoskedastic_model_learnable_noise.parameters()) [Parameter containing:
tensor(0., device='cuda:0', requires_grad=True),
Parameter containing:
tensor([[-0.7584],
[-0.9339],
[ 0.0176],
[ 0.9118]], device='cuda:0', requires_grad=True),
Parameter containing:
tensor([ 0.5769, -0.5822, -0.1298, -0.7372], device='cuda:0',
requires_grad=True),
Parameter containing:
tensor([[-0.2412, 0.0905, 0.2723, 0.4142],
[-0.4591, 0.3343, -0.3526, 0.1872],
[ 0.4231, 0.0070, 0.4549, -0.4260],
[-0.1910, 0.2916, -0.1089, -0.1024]], device='cuda:0',
requires_grad=True),
Parameter containing:
tensor([-0.2084, 0.3447, 0.2453, 0.1602], device='cuda:0',
requires_grad=True),
Parameter containing:
tensor([[-0.2810, -0.4059, 0.0541, 0.1481]], device='cuda:0',
requires_grad=True),
Parameter containing:
tensor([-0.2309], device='cuda:0', requires_grad=True)]jtu.tree_leaves(params) == [log_noise_std] + list(homoskedastic_model_learnable_noise.parameters()) True
train_fn(
homoskedastic_model_learnable_noise,
loss_homoskedastic_noise,
params,
x_lin[:, None],
y,
n_epochs=1000,
lr=0.01,
)Epoch 0: loss 0.948
Epoch 50: loss 0.481
Epoch 100: loss 0.102
Epoch 150: loss -0.108
Epoch 200: loss -0.156
Epoch 250: loss -0.158
Epoch 300: loss -0.158
Epoch 350: loss -0.158
Epoch 400: loss -0.158
Epoch 450: loss -0.158
Epoch 500: loss -0.158
Epoch 550: loss -0.158
Epoch 600: loss -0.158
Epoch 650: loss -0.158
Epoch 700: loss -0.158
Epoch 750: loss -0.158
Epoch 800: loss -0.158
Epoch 850: loss -0.158
Epoch 900: loss -0.158
Epoch 950: loss -0.158-0.15807949006557465# Plot the trained model
with torch.no_grad():
y_hat = homoskedastic_model_learnable_noise(x_lin[:, None]).squeeze()
aleatoric_std = torch.exp(log_noise_std)
plot_results(y_hat, aleatoric_std=aleatoric_std, model_name="Trained model")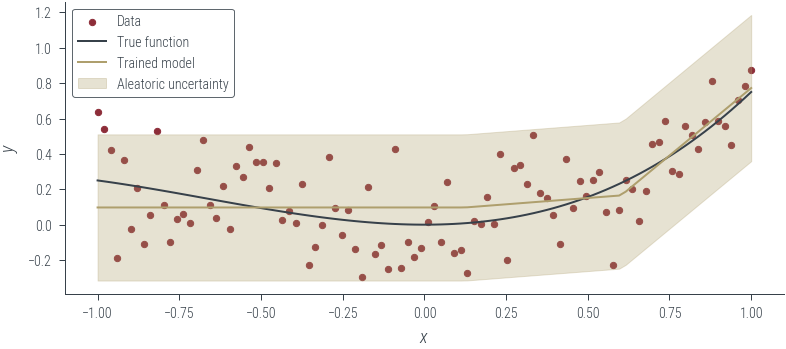
Case 1.2: Models assuming heteroskedastic noise
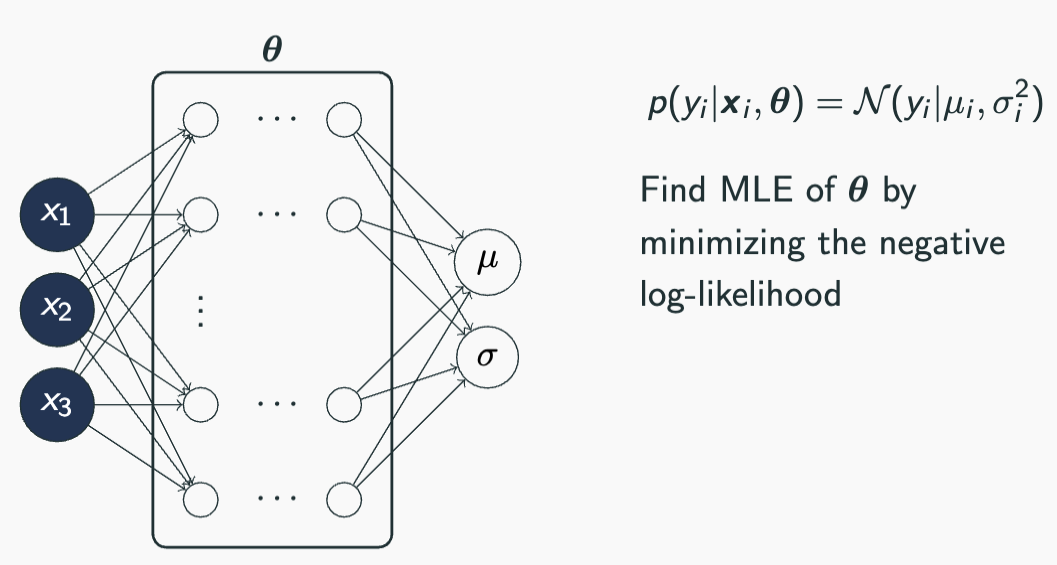
class HeteroskedasticNN(torch.nn.Module):
def __init__(self, n_hidden=10):
super().__init__()
self.fc1 = torch.nn.Linear(1, n_hidden)
self.fc2 = torch.nn.Linear(n_hidden, n_hidden)
self.fc3 = torch.nn.Linear(n_hidden, 2) # we learn both mu and log_noise_std
def forward(self, x):
x = self.fc1(x)
x = torch.relu(x)
x = self.fc2(x)
x = torch.relu(x)
z = self.fc3(x)
mu_hat = z[:, 0]
log_noise_std = z[:, 1]
return mu_hat, log_noise_stdheteroskedastic_model = HeteroskedasticNN().to(device)
heteroskedastic_modelHeteroskedasticNN(
(fc1): Linear(in_features=1, out_features=10, bias=True)
(fc2): Linear(in_features=10, out_features=10, bias=True)
(fc3): Linear(in_features=10, out_features=2, bias=True)
)# Plot the untrained model
with torch.no_grad():
y_hat, log_noise_std = heteroskedastic_model(x_lin[:, None])
aleatoric_std = torch.exp(log_noise_std)
plot_results(y_hat, aleatoric_std=aleatoric_std, model_name="Untrained model")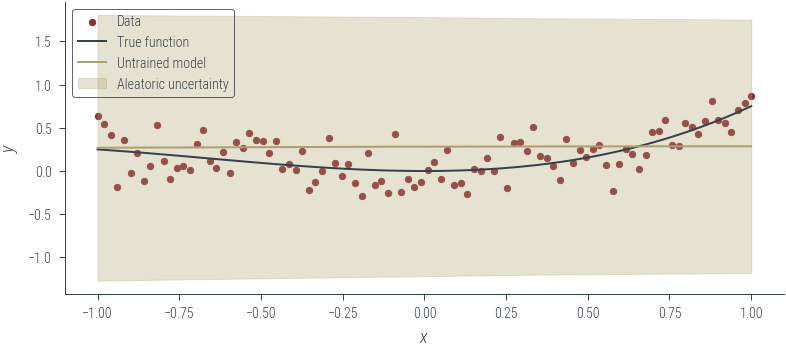
def loss_heteroskedastic(model, x, y, params):
mu_hat, log_noise_std = model(x)
noise_std = torch.exp(log_noise_std)
dist = torch.distributions.Normal(mu_hat, noise_std)
return -dist.log_prob(y).mean()
params = list(heteroskedastic_model.parameters())
train_fn(
heteroskedastic_model,
loss_heteroskedastic,
params,
x_lin[:, None],
y,
n_epochs=1000,
lr=0.01,
)Epoch 0: loss 0.702
Epoch 50: loss -0.130
Epoch 100: loss -0.230
Epoch 150: loss -0.250
Epoch 200: loss -0.256
Epoch 250: loss -0.260
Epoch 300: loss -0.262
Epoch 350: loss -0.264
Epoch 400: loss -0.266
Epoch 450: loss -0.266
Epoch 500: loss -0.267
Epoch 550: loss -0.267
Epoch 600: loss -0.267
Epoch 650: loss -0.267
Epoch 700: loss -0.267
Epoch 750: loss -0.267
Epoch 800: loss -0.267
Epoch 850: loss -0.267
Epoch 900: loss -0.267
Epoch 950: loss -0.267-0.2661149203777313# Plot the trained model
with torch.no_grad():
y_hat, log_noise_std = heteroskedastic_model(x_lin[:, None])
aleatoric_std = torch.exp(log_noise_std)
plot_results(y_hat, aleatoric_std=aleatoric_std, model_name="Trained model")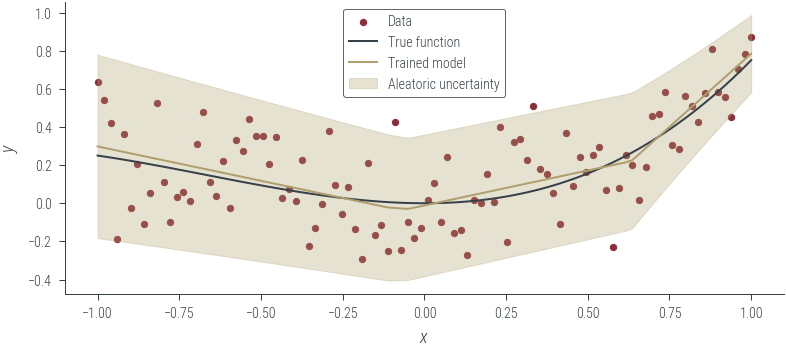
Data with heteroskedastic noise
torch.manual_seed(42)
N = 100
x_lin = torch.linspace(-1, 1, N)
f = lambda x: 0.5 * x**2 + 0.25 * x**3
eps = torch.randn(N) * (0.1 + 0.4 * x_lin)
y = f(x_lin) + eps
# Move to GPU
x_lin = x_lin.to(device)
y = y.to(device)
# Plot data and true function
plt.plot(x_lin.cpu(), y.cpu(), "o", label="data")
plt.plot(x_lin.cpu(), f(x_lin).cpu(), label="true function")
plt.xlabel("x")
plt.ylabel("y")
plt.legend()<matplotlib.legend.Legend at 0x7f5ea8af9df0>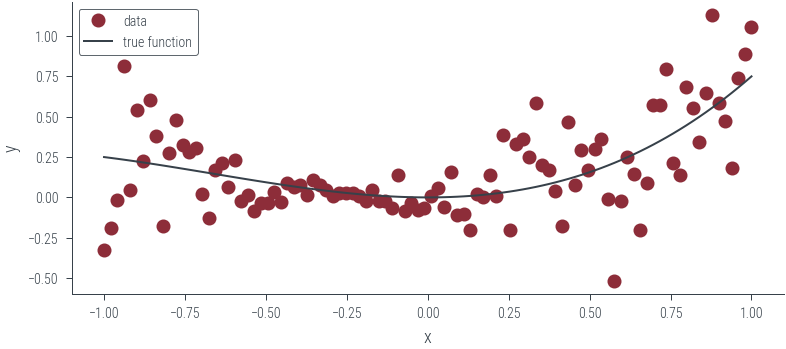
homoskedastic_model_fixed_noise = MeanEstimateNN().to(device)
fixed_log_noise_std = torch.log(torch.tensor(0.5)).to(device)
# Plot the untrained model
with torch.no_grad():
y_hat = homoskedastic_model_fixed_noise(x_lin[:, None]).squeeze()
aleatoric_std = torch.exp(fixed_log_noise_std)
plot_results(y_hat, aleatoric_std=aleatoric_std, model_name="Untrained model\n (Homosdedastic fixed noise)")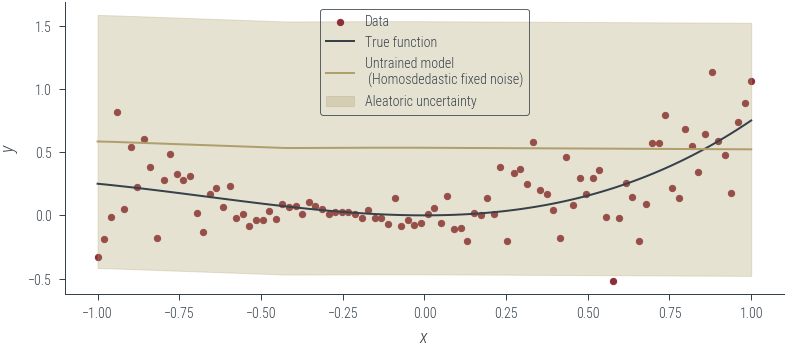
params = {
"nn_params": list(homoskedastic_model_fixed_noise.parameters()),
"log_noise_std": fixed_log_noise_std,
}
train_fn(
homoskedastic_model_fixed_noise,
loss_homoskedastic_noise,
params,
x_lin[:, None],
y,
n_epochs=1000,
lr=0.001,
)
# Plot the trained model
with torch.no_grad():
y_hat = homoskedastic_model_fixed_noise(x_lin[:, None]).squeeze()
aleatoric_std = torch.exp(fixed_log_noise_std)
plot_results(y_hat, aleatoric_std=aleatoric_std, model_name="Trained model\n (Homosdedastic fixed noise)")Epoch 0: loss 0.668
Epoch 50: loss 0.543
Epoch 100: loss 0.466
Epoch 150: loss 0.423
Epoch 200: loss 0.401
Epoch 250: loss 0.393
Epoch 300: loss 0.389
Epoch 350: loss 0.385
Epoch 400: loss 0.382
Epoch 450: loss 0.379
Epoch 500: loss 0.376
Epoch 550: loss 0.372
Epoch 600: loss 0.369
Epoch 650: loss 0.366
Epoch 700: loss 0.362
Epoch 750: loss 0.358
Epoch 800: loss 0.354
Epoch 850: loss 0.350
Epoch 900: loss 0.347
Epoch 950: loss 0.344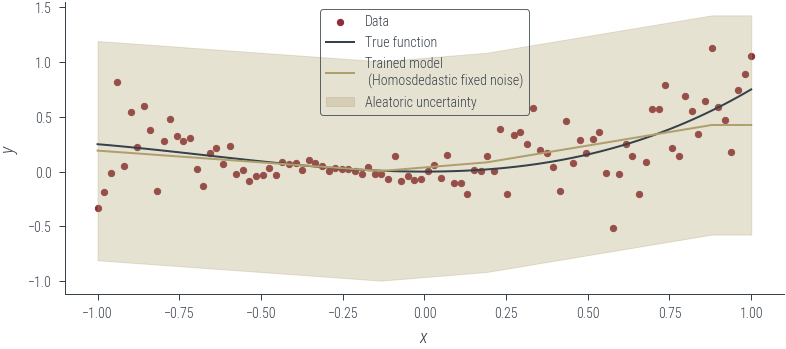
# Now, fit the homoskedastic model with learned noise
homoskedastic_model_learnable_noise = MeanEstimateNN().to(device)
log_noise_std = torch.nn.Parameter(torch.tensor(0.0).to(device))
# Plot the untrained model
with torch.no_grad():
y_hat = homoskedastic_model_learnable_noise(x_lin[:, None]).squeeze()
aleatoric_std = torch.exp(log_noise_std)
plot_results(y_hat, aleatoric_std=aleatoric_std, model_name="Untrained model\n (Homosdedastic learnable noise)")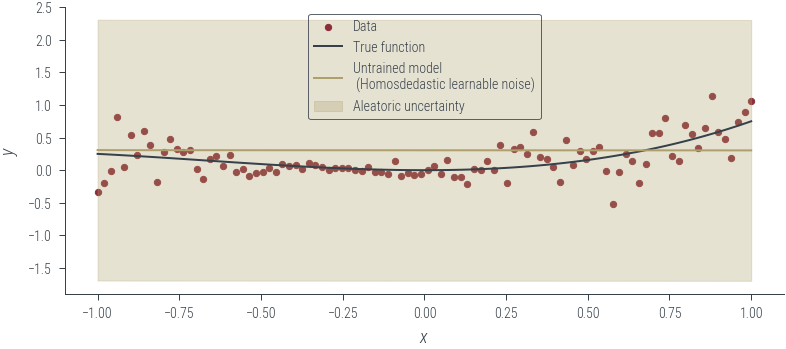
# Train the model
params = {
"nn_params": list(homoskedastic_model_learnable_noise.parameters()),
"log_noise_std": log_noise_std,
}
train_fn(
homoskedastic_model_learnable_noise,
loss_homoskedastic_noise,
params,
x_lin[:, None],
y,
n_epochs=1000,
lr=0.01,
)
# Plot the trained model
with torch.no_grad():
y_hat = homoskedastic_model_learnable_noise(x_lin[:, None]).squeeze()
aleatoric_std = torch.exp(log_noise_std)
plot_results(y_hat, aleatoric_std=aleatoric_std, model_name="Trained model\n (Homosdedastic fixed noise)")Epoch 0: loss 0.971
Epoch 50: loss 0.523
Epoch 100: loss 0.143
Epoch 150: loss -0.036
Epoch 200: loss -0.063
Epoch 250: loss -0.063
Epoch 300: loss -0.063
Epoch 350: loss -0.063
Epoch 400: loss -0.063
Epoch 450: loss -0.063
Epoch 500: loss -0.063
Epoch 550: loss -0.063
Epoch 600: loss -0.063
Epoch 650: loss -0.063
Epoch 700: loss -0.063
Epoch 750: loss -0.063
Epoch 800: loss -0.063
Epoch 850: loss -0.063
Epoch 900: loss -0.063
Epoch 950: loss -0.063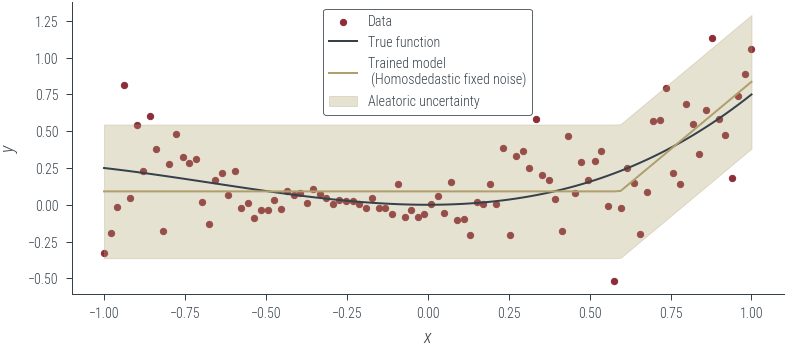
# Now, fit the heteroskedastic model
heteroskedastic_model = HeteroskedasticNN().to(device)
# Plot the untrained model
with torch.no_grad():
y_hat, log_noise_std = heteroskedastic_model(x_lin[:, None])
aleatoric_std = torch.exp(log_noise_std)
plot_results(y_hat, aleatoric_std=aleatoric_std, model_name="Untrained model\n (Heteroskedastic)")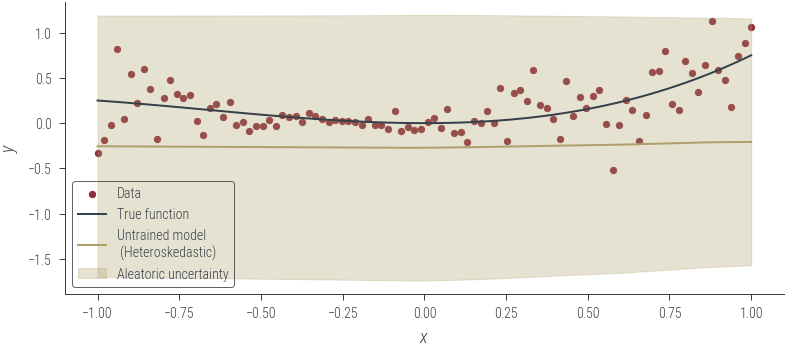
# Train the model
params = list(heteroskedastic_model.parameters())
train_fn(
heteroskedastic_model,
loss_heteroskedastic,
params,
x_lin[:, None],
y,
n_epochs=1000,
lr=0.01,
)Epoch 0: loss 0.849
Epoch 50: loss 0.023
Epoch 100: loss -0.083
Epoch 150: loss -0.334
Epoch 200: loss -0.446
Epoch 250: loss -0.476
Epoch 300: loss -0.488
Epoch 350: loss -0.508
Epoch 400: loss -0.536
Epoch 450: loss -0.547
Epoch 500: loss -0.558
Epoch 550: loss -0.565
Epoch 600: loss -0.570
Epoch 650: loss -0.569
Epoch 700: loss -0.574
Epoch 750: loss -0.577
Epoch 800: loss -0.586
Epoch 850: loss -0.589
Epoch 900: loss -0.597
Epoch 950: loss -0.602-0.6076287031173706# Plot the trained model
with torch.no_grad():
y_hat, log_noise_std = heteroskedastic_model(x_lin[:, None])
aleatoric_std = torch.exp(log_noise_std)
plot_results(y_hat, aleatoric_std=aleatoric_std, model_name="Trained model\n (Heteroskedastic)")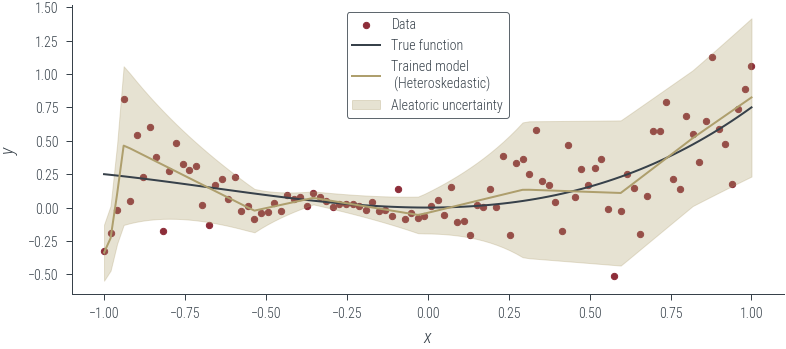
Epistemic Uncertainty: Bayesian NN with Laplace approximation
MAP estimation
def negative_log_prior(model):
log_prior = 0.0
for param in model.parameters():
log_prior += torch.distributions.Normal(0, 1).log_prob(param).sum()
return -log_prior
def negative_log_likelihood(model, x, y, log_noise_std):
mu_hat = model(x).squeeze()
assert mu_hat.shape == y.shape
noise_std = torch.exp(log_noise_std).expand_as(mu_hat)
dist = torch.distributions.Normal(mu_hat, noise_std)
return -dist.log_prob(y).sum()
def negative_log_joint(model, x, y, log_noise_std):
return negative_log_likelihood(model, x, y, log_noise_std) + negative_log_prior(
model
)def custom_loss_fn(model, x, y, params):
log_noise_std = params["log_noise_std"]
return negative_log_joint(model, x, y, log_noise_std)
torch.manual_seed(3)
laplace_model = MeanEstimateNN().to(device)
fixed_log_noise_std = torch.log(torch.tensor(0.2)).to(device)
params = {
"log_noise_std": fixed_log_noise_std,
"nn_params": list(laplace_model.parameters()),
}
train_fn(
laplace_model,
custom_loss_fn,
params,
x_lin[:, None],
y,
n_epochs=1000,
lr=0.01,
)Epoch 0: loss 439.902
Epoch 50: loss 138.376
Epoch 100: loss 131.914
Epoch 150: loss 131.173
Epoch 200: loss 130.548
Epoch 250: loss 126.990
Epoch 300: loss 125.752
Epoch 350: loss 125.163
Epoch 400: loss 123.350
Epoch 450: loss 122.710
Epoch 500: loss 122.472
Epoch 550: loss 122.251
Epoch 600: loss 122.128
Epoch 650: loss 122.113
Epoch 700: loss 121.922
Epoch 750: loss 121.868
Epoch 800: loss 121.792
Epoch 850: loss 121.794
Epoch 900: loss 121.696
Epoch 950: loss 121.726121.6390151977539with torch.no_grad():
y_hat = laplace_model(x_lin[:, None]).squeeze()
aleatoric_std = torch.exp(fixed_log_noise_std)
plot_results(y_hat, aleatoric_std=aleatoric_std, model_name="MAP estimate\n (Homosdedastic fixed noise)")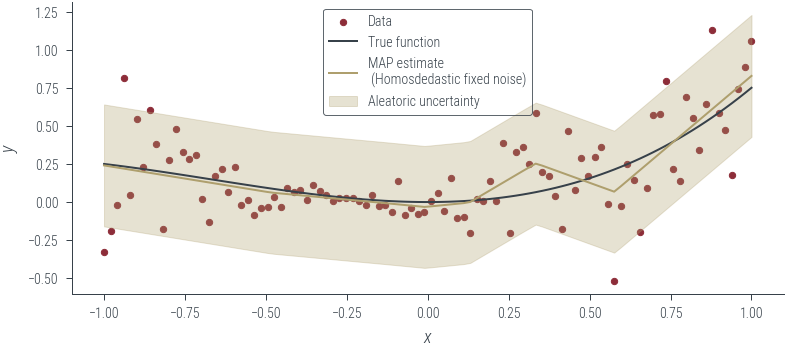
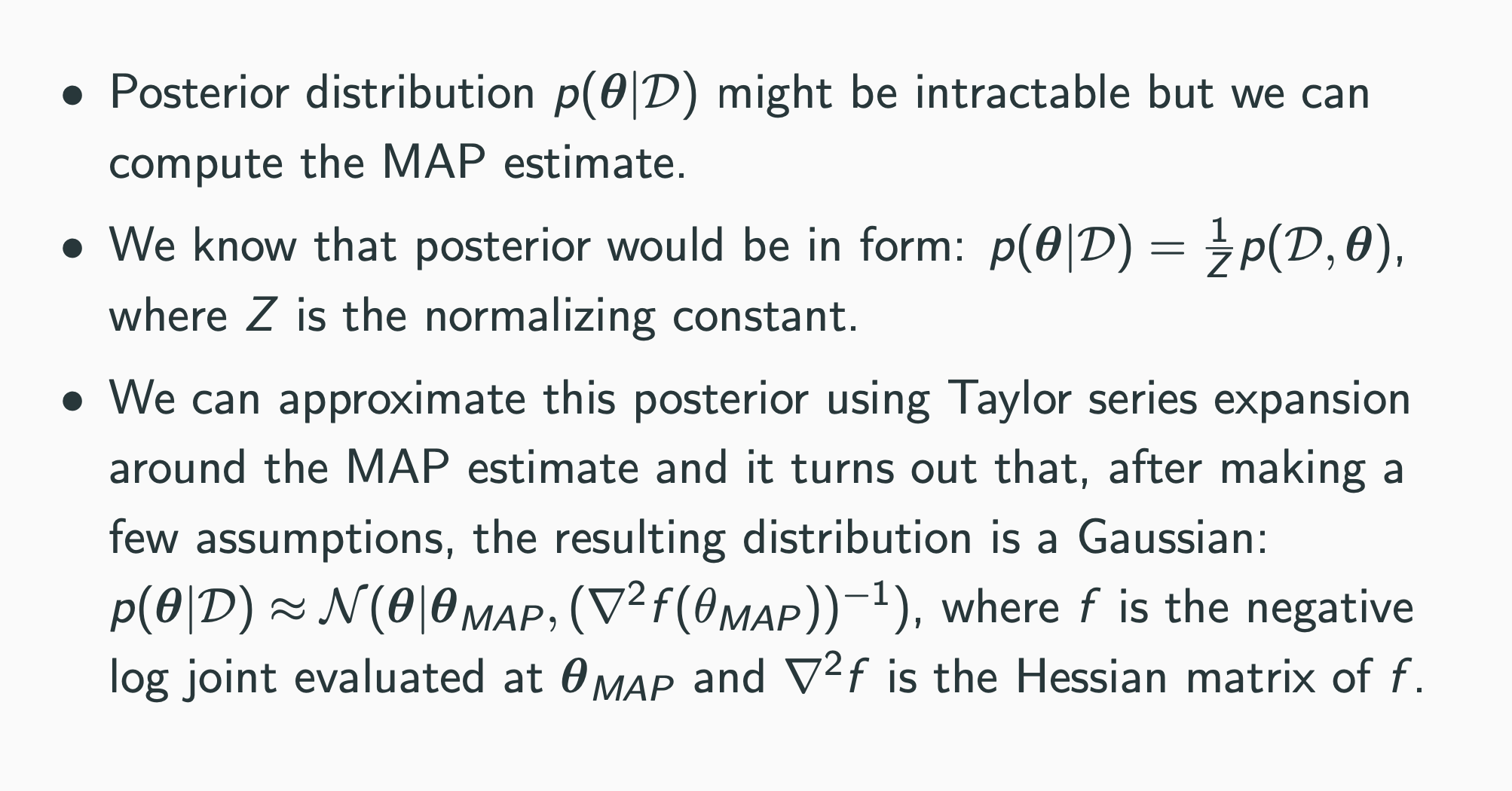
What weighs to consider?
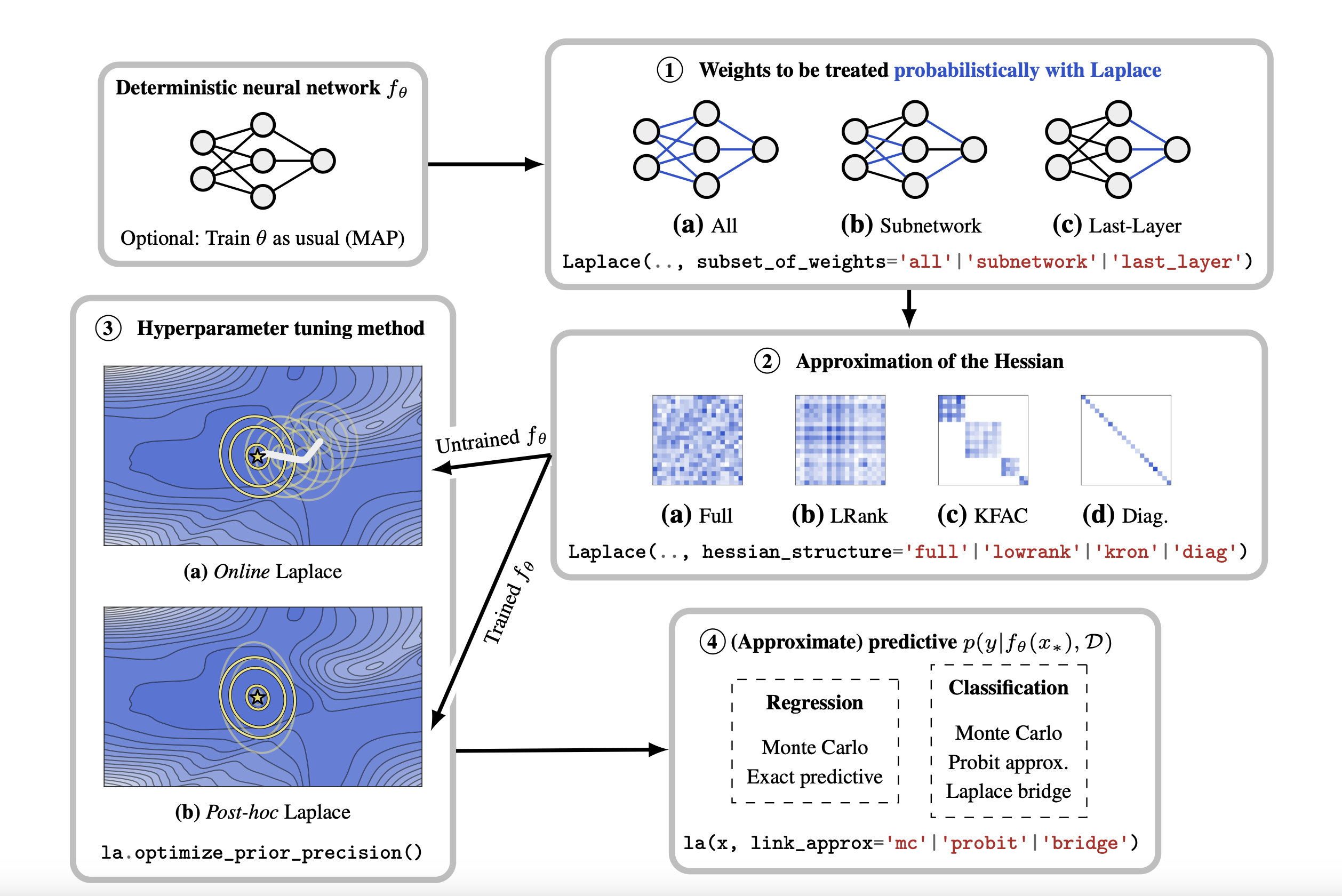
Goal: Compute the Hessian of the negative log joint wrt the last layer weights
Challenge: The negative log joint is a function of all the weights, not just the last layer weights
Aside on functools.partial
The functools module is for higher-order functions: functions that act on or return other functions. In general, any callable object can be treated as a function for the purposes of this module.
print(int("1001", base=2), int("1001", base=4), int("1001"))
from functools import partial
base_two = partial(int, base=2)
base_two.__doc__ = "Convert base 2 string to an int."
print(base_two)
print(base_two.__doc__)
print(help(base_two))
print(base_two("1001"))9 65 1001
functools.partial(<class 'int'>, base=2)
Convert base 2 string to an int.
Help on partial:
functools.partial(<class 'int'>, base=2)
Convert base 2 string to an int.
None
9A primer on functional calls to PyTorch
tiny_model = torch.nn.Linear(3, 1)
input = torch.randn(2, 3)
target = torch.randn(2, 1)
tiny_modelLinear(in_features=3, out_features=1, bias=True)output = tiny_model(input)
outputtensor([[ 0.0756],
[-0.2420]], grad_fn=<AddmmBackward0>)loss_fn = torch.nn.MSELoss()
loss = loss_fn(output, target)
loss.backward()
grad_dict = {"weight": tiny_model.weight.grad, "bias": tiny_model.bias.grad}
grad_dict{'weight': tensor([[ 2.2469, -1.7073, -0.6623]]), 'bias': tensor([-1.9448])}params = dict(tiny_model.named_parameters())
output = torch.func.functional_call(tiny_model, params, input)
outputtensor([[ 0.0756],
[-0.2420]], grad_fn=<AddmmBackward0>)def custom_loss_fn(params):
output = torch.func.functional_call(tiny_model, params, input)
return loss_fn(output, target)
torch.func.grad(custom_loss_fn, argnums=0)(params){'weight': tensor([[ 2.2469, -1.7073, -0.6623]], grad_fn=<TBackward0>),
'bias': tensor([-1.9448], grad_fn=<ViewBackward0>)}It is also possible to get the gradients/hessian with respect to only a few weights.
def custom_loss_fn(partial_params, params):
params.update(partial_params)
output = torch.func.functional_call(tiny_model, params, input)
return loss_fn(output, target)
partial_params = {"bias": params["bias"]}
torch.func.grad(custom_loss_fn, argnums=0)(partial_params, params){'bias': tensor([-1.9448], grad_fn=<ViewBackward0>)}Last layer Laplace approximation
def functional_negative_log_prior(partial_params):
partial_parameter_leaves = jtu.tree_leaves(partial_params)
log_prior = 0.0
for param in partial_parameter_leaves:
log_prior += torch.distributions.Normal(0, 1).log_prob(param).sum()
return -log_prior
def functional_negative_log_likelihood(
partial_params, params, model, x, y, log_noise_std
):
params.update(partial_params)
mu_hat = torch.func.functional_call(model, params, x).squeeze()
assert mu_hat.shape == y.shape, f"{mu_hat.shape} != {y.shape}"
noise_std = torch.exp(log_noise_std).expand_as(mu_hat)
dist = torch.distributions.Normal(mu_hat, noise_std)
return -dist.log_prob(y).sum()
def functional_negative_log_joint(partial_params, params, model, x, y, log_noise_std):
return functional_negative_log_likelihood(
partial_params, params, model, x, y, log_noise_std
) + functional_negative_log_prior(partial_params)params = dict(laplace_model.named_parameters())
params.keys()dict_keys(['fc1.weight', 'fc1.bias', 'fc2.weight', 'fc2.bias', 'fc3.weight', 'fc3.bias'])partial_params = {"fc3.weight": params["fc3.weight"]}
partial_params{'fc3.weight': Parameter containing:
tensor([[ 1.3786e-12, 3.2083e-01, 8.6144e-01, 2.3610e-03, -1.3280e+00,
1.9577e-01, 8.8147e-01, 2.7229e-01, -4.8347e-05, 2.0937e-01]],
device='cuda:0', requires_grad=True)}jtu.tree_leaves(partial_params)[Parameter containing:
tensor([[ 1.3786e-12, 3.2083e-01, 8.6144e-01, 2.3610e-03, -1.3280e+00,
1.9577e-01, 8.8147e-01, 2.7229e-01, -4.8347e-05, 2.0937e-01]],
device='cuda:0', requires_grad=True)]print("Full negative log prior", negative_log_prior(laplace_model))
print(
"Partial negative log prior",
functional_negative_log_prior(partial_params),
)Full negative log prior tensor(135.0255, device='cuda:0', grad_fn=<NegBackward0>)
Partial negative log prior tensor(10.9603, device='cuda:0', grad_fn=<NegBackward0>)print(
"Full negative log likelihood",
negative_log_likelihood(laplace_model, x_lin[:, None], y, fixed_log_noise_std),
)
print(
"Partial negative log likelihood",
functional_negative_log_likelihood(
partial_params, params, laplace_model, x_lin[:, None], y, fixed_log_noise_std
),
)Full negative log likelihood tensor(-13.3980, device='cuda:0', grad_fn=<NegBackward0>)
Partial negative log likelihood tensor(-13.3980, device='cuda:0', grad_fn=<NegBackward0>)print(
"Full negative log joint",
negative_log_joint(laplace_model, x_lin[:, None], y, fixed_log_noise_std),
)
print(
"Partial negative log joint",
functional_negative_log_joint(
partial_params, params, laplace_model, x_lin[:, None], y, fixed_log_noise_std
),
)Full negative log joint tensor(121.6275, device='cuda:0', grad_fn=<AddBackward0>)
Partial negative log joint tensor(-2.4377, device='cuda:0', grad_fn=<AddBackward0>)map_params = dict(laplace_model.named_parameters())
last_layer_params = {"fc3.weight": map_params["fc3.weight"]}
partial_func = partial(
functional_negative_log_joint,
params=map_params,
model=laplace_model,
x=x_lin[:, None],
y=y,
log_noise_std=fixed_log_noise_std,
)
H = torch.func.hessian(partial_func)(last_layer_params)["fc3.weight"]["fc3.weight"]
print(H.shape)
H = H[0, :, 0, :]
print(H.shape)torch.Size([1, 10, 1, 10])
torch.Size([10, 10])import seaborn as sns
with torch.no_grad():
cov = torch.inverse(H + 1e-3 * torch.eye(H.shape[0]).to(device))
plt.figure(figsize=(7, 7))
sns.heatmap(cov.cpu().numpy(), annot=True, fmt=".2f", cmap="viridis")
plt.gca().set_aspect("equal", "box")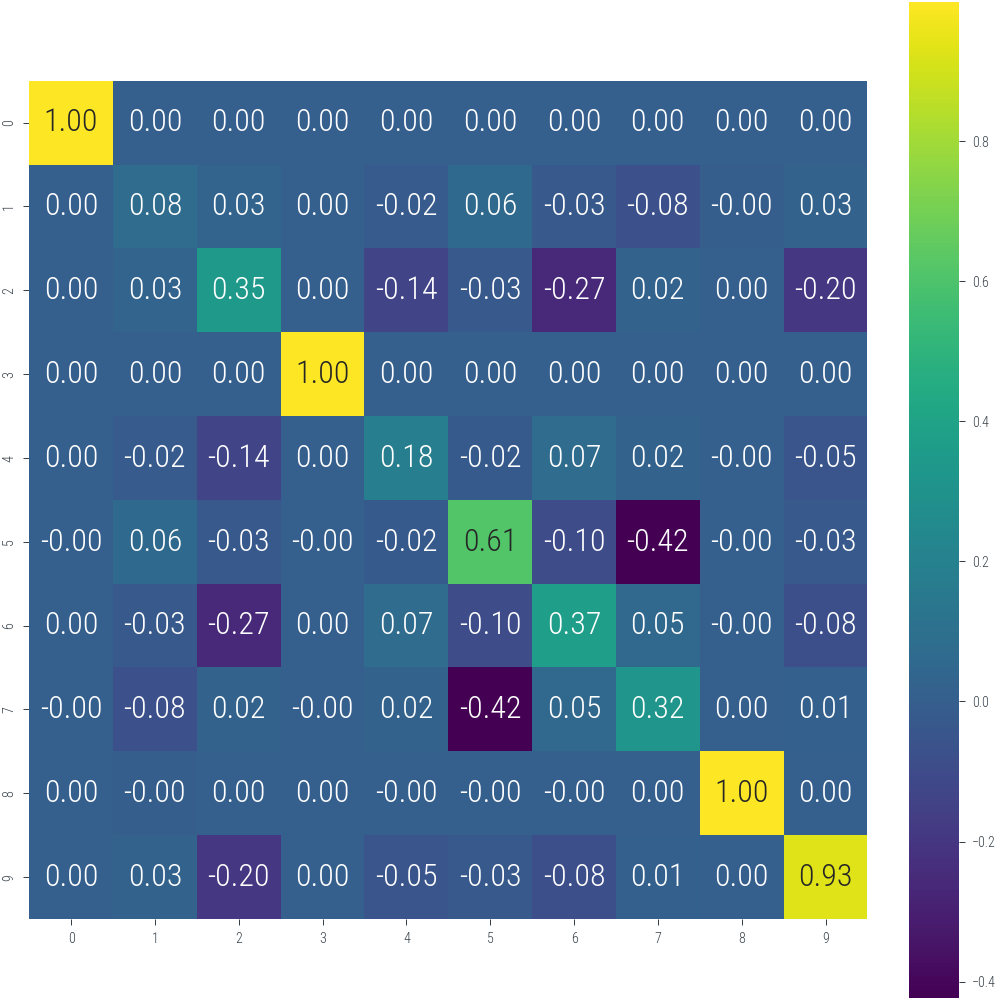
laplace_posterior = torch.distributions.MultivariateNormal(
last_layer_params["fc3.weight"].ravel(), cov
)
last_layer_weights_samples = laplace_posterior.sample((501,))[..., None]
last_layer_weights_samples.shapetorch.Size([501, 10, 1])def forward_pass(last_layer_weight, params):
params.update({"fc3.weight": last_layer_weight.reshape(1, -1)})
return torch.func.functional_call(laplace_model, params, x_lin[:, None]).squeeze()
forward_pass(last_layer_weights_samples[0], params).shapetorch.Size([100])mc_outputs = torch.vmap(lambda x: forward_pass(x, params))(last_layer_weights_samples)
print(mc_outputs.shape)torch.Size([501, 100])mean_mc_outputs = mc_outputs.mean(0)
std_mc_outputs = mc_outputs.std(0)
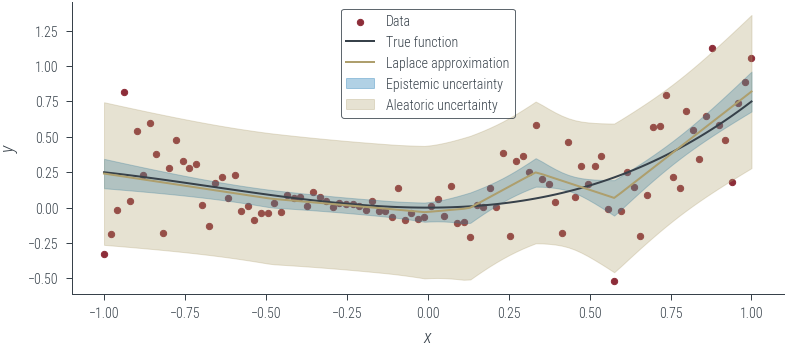
mean_mc_outputs.shape, std_mc_outputs.shape(torch.Size([100]), torch.Size([100]))with torch.no_grad():
epistemic_std = std_mc_outputs
aleatoric_std = torch.exp(fixed_log_noise_std) + epistemic_std
plot_results(
mean_mc_outputs,
epistemic_std=epistemic_std,
aleatoric_std=aleatoric_std,
model_name="Laplace approximation",
)
with torch.no_grad():
epistemic_std = std_mc_outputs
aleatoric_std = torch.exp(fixed_log_noise_std) + epistemic_std
plot_results(
mean_mc_outputs,
epistemic_std=None,
aleatoric_std=None,
model_name="Laplace approximation",
)
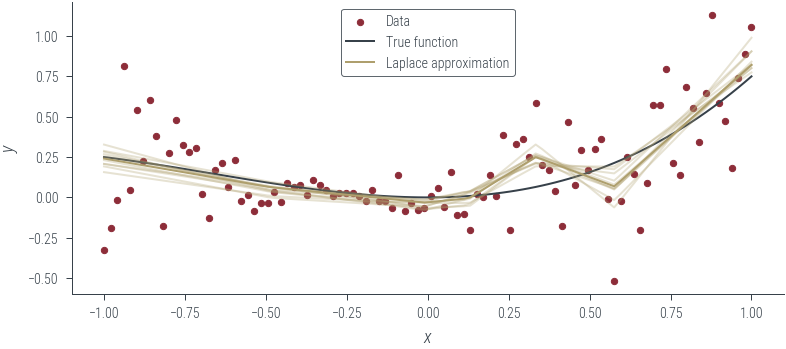
for i in range(10):
plt.plot(
x_lin.cpu(),
mc_outputs[i].cpu(),
alpha=0.3,
color="C2",
label="Laplace approximation",
)