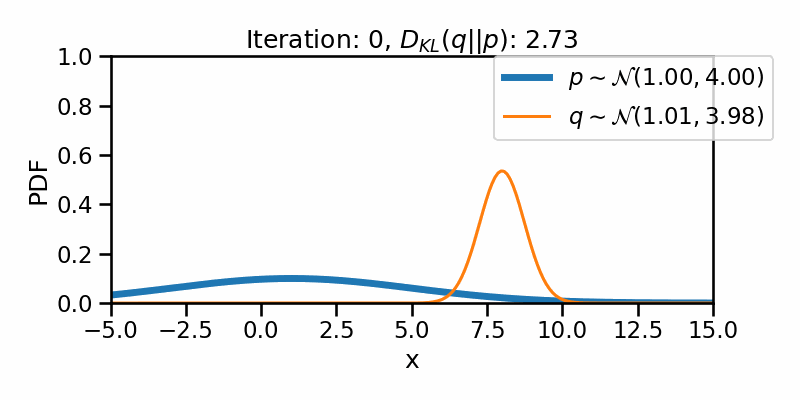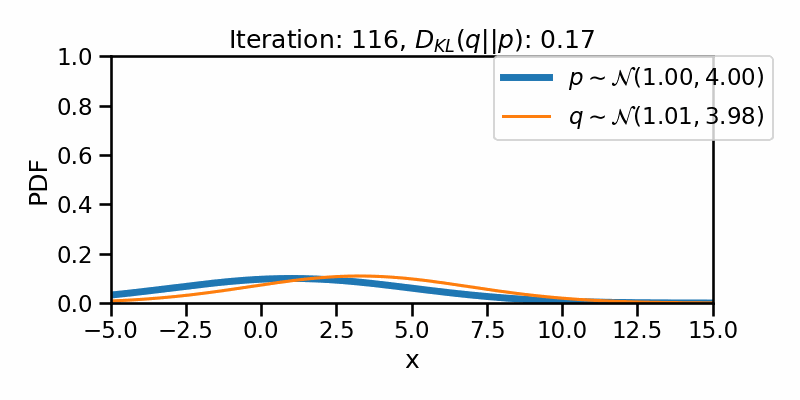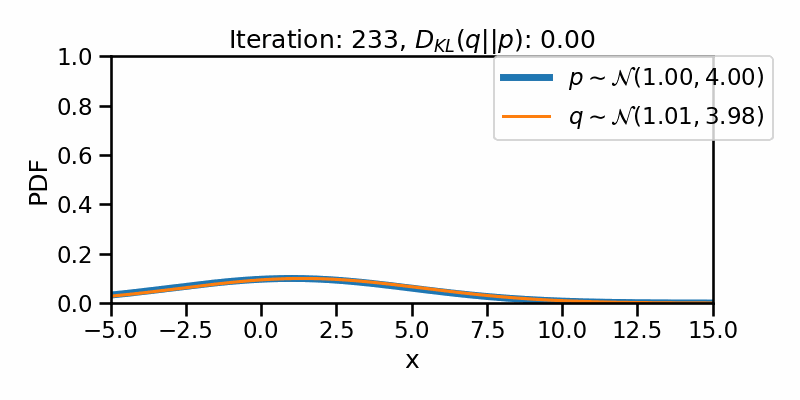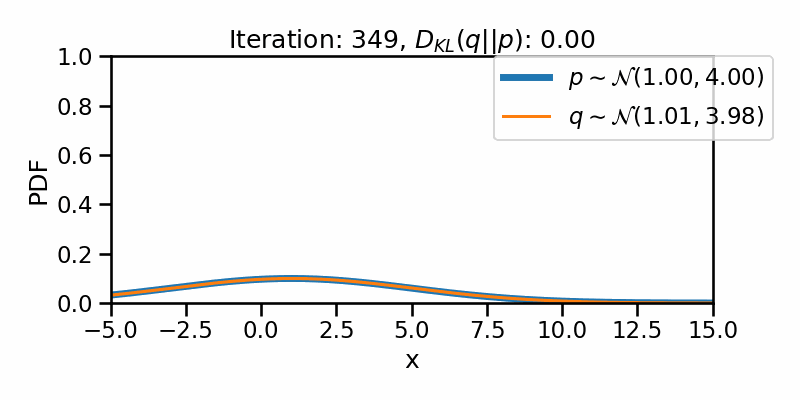
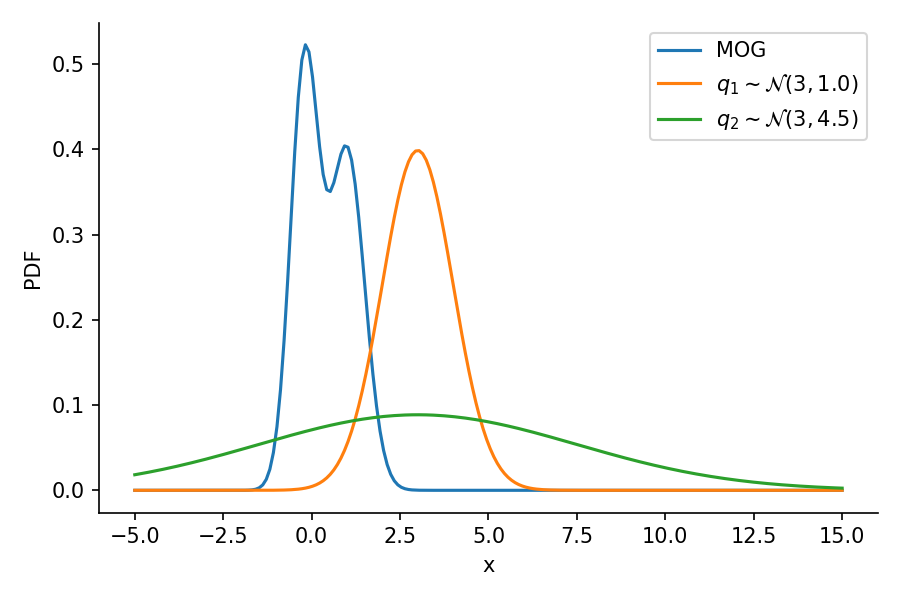
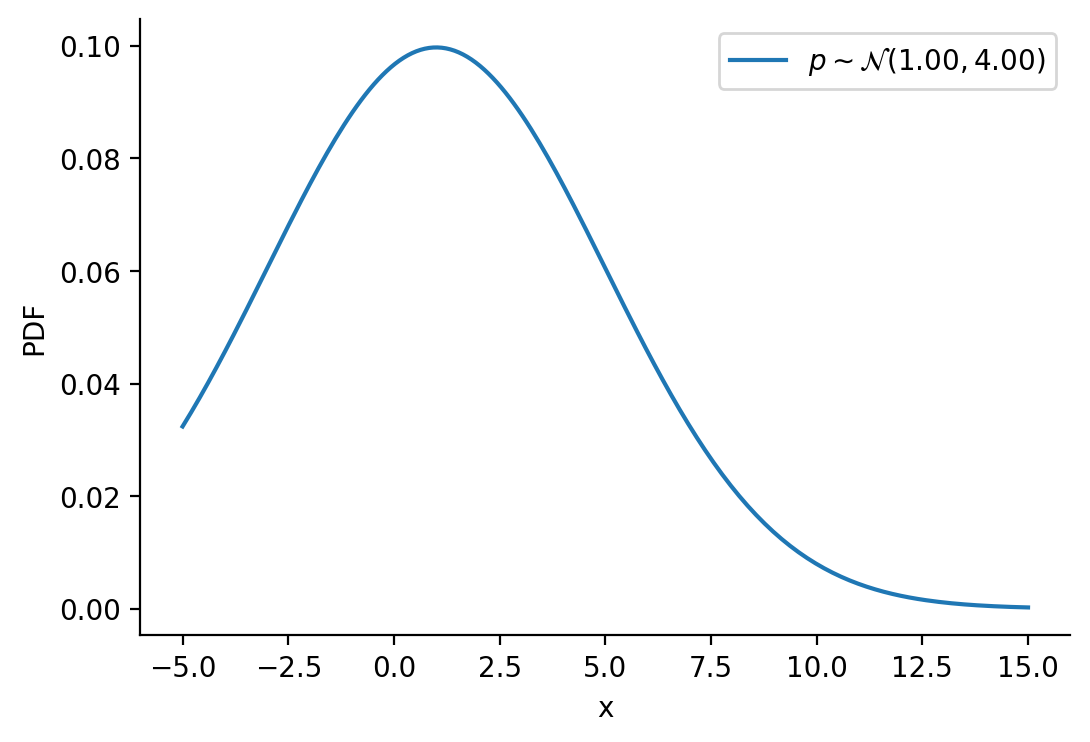
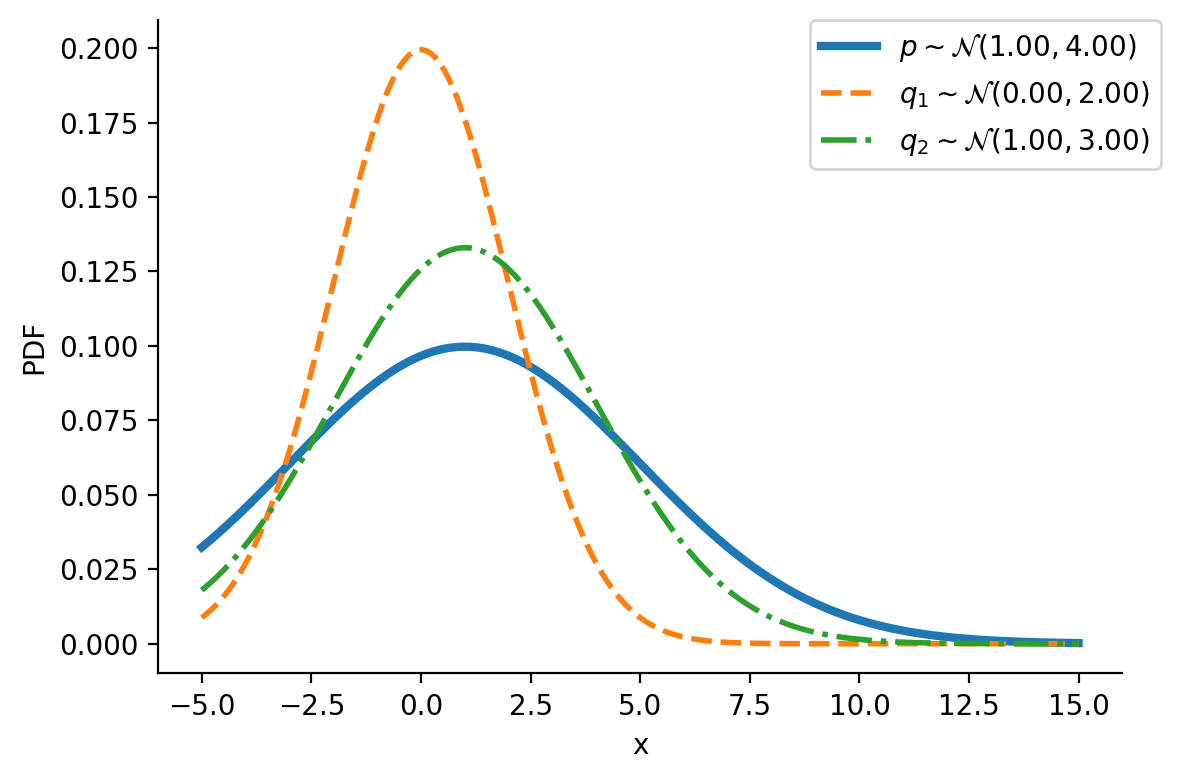
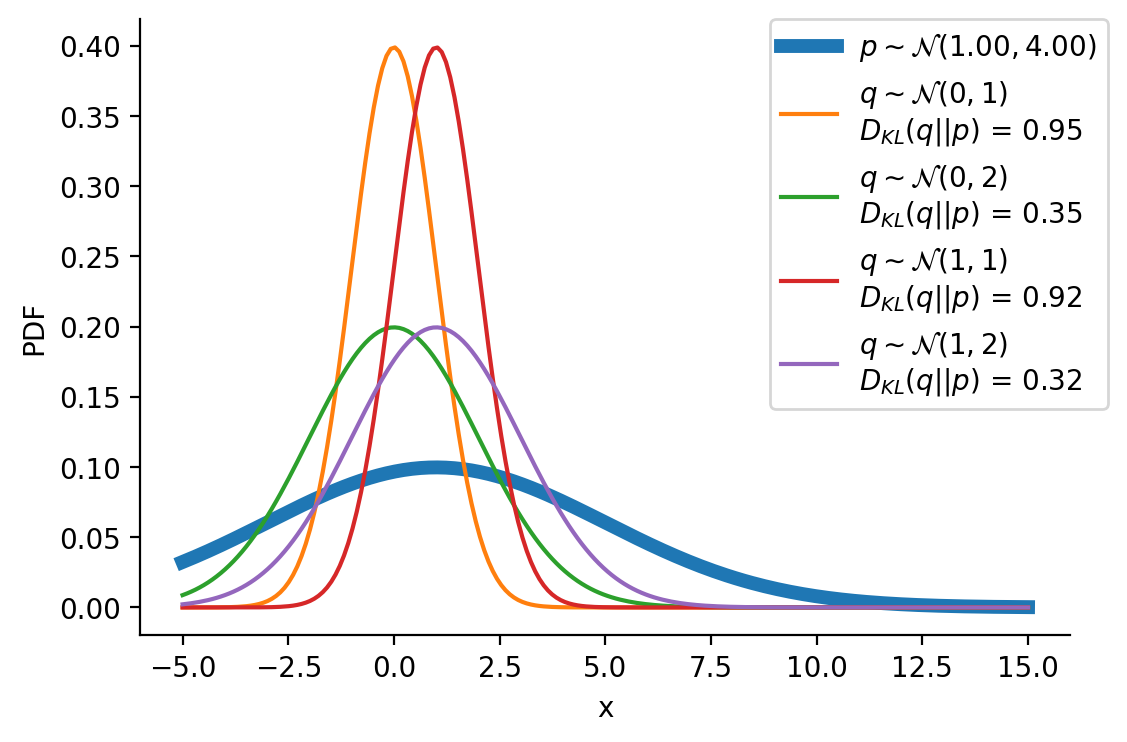
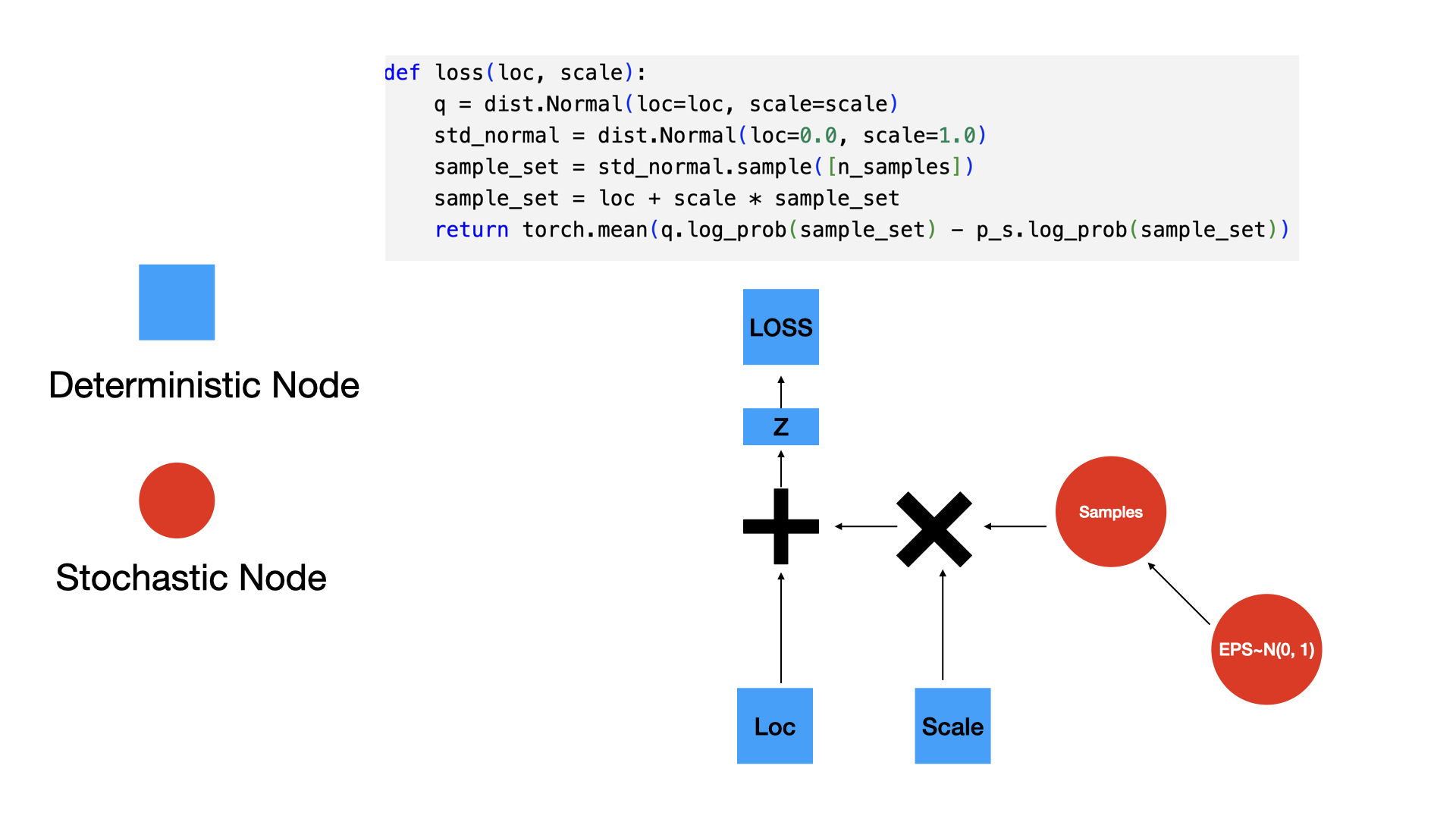
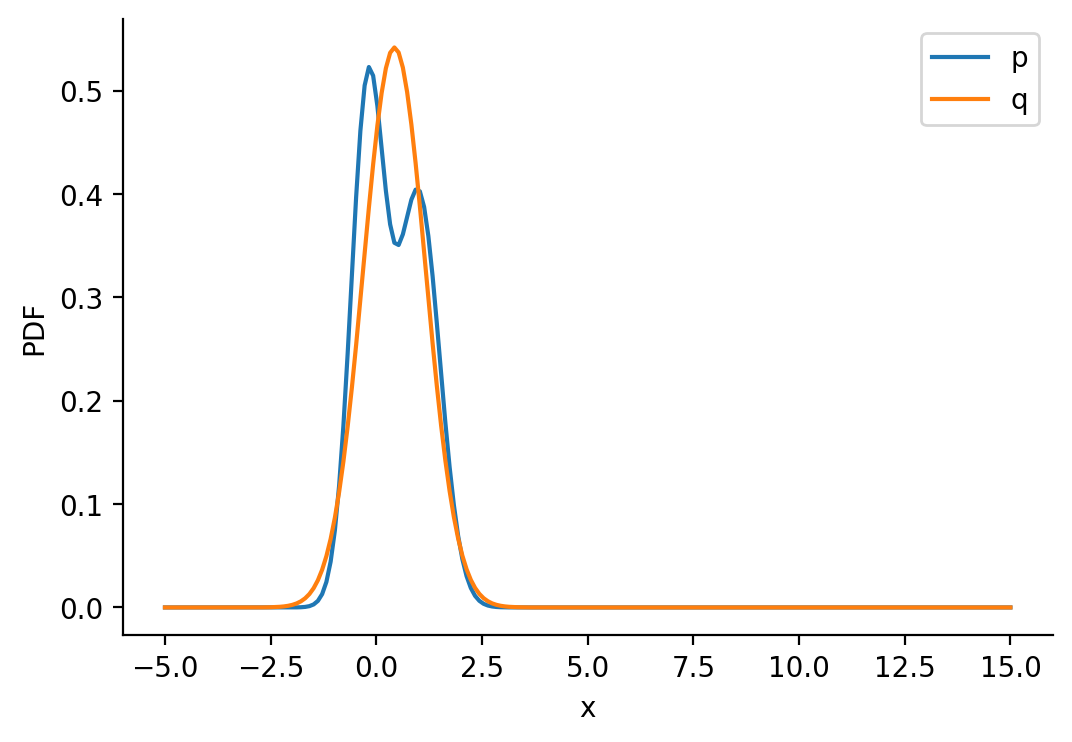
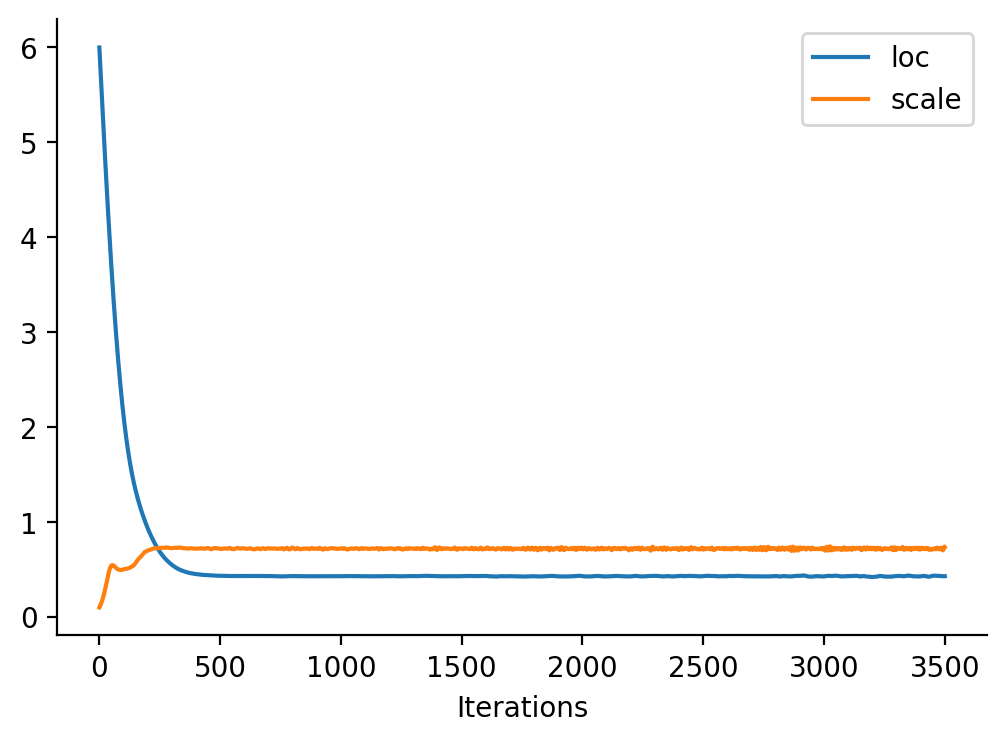
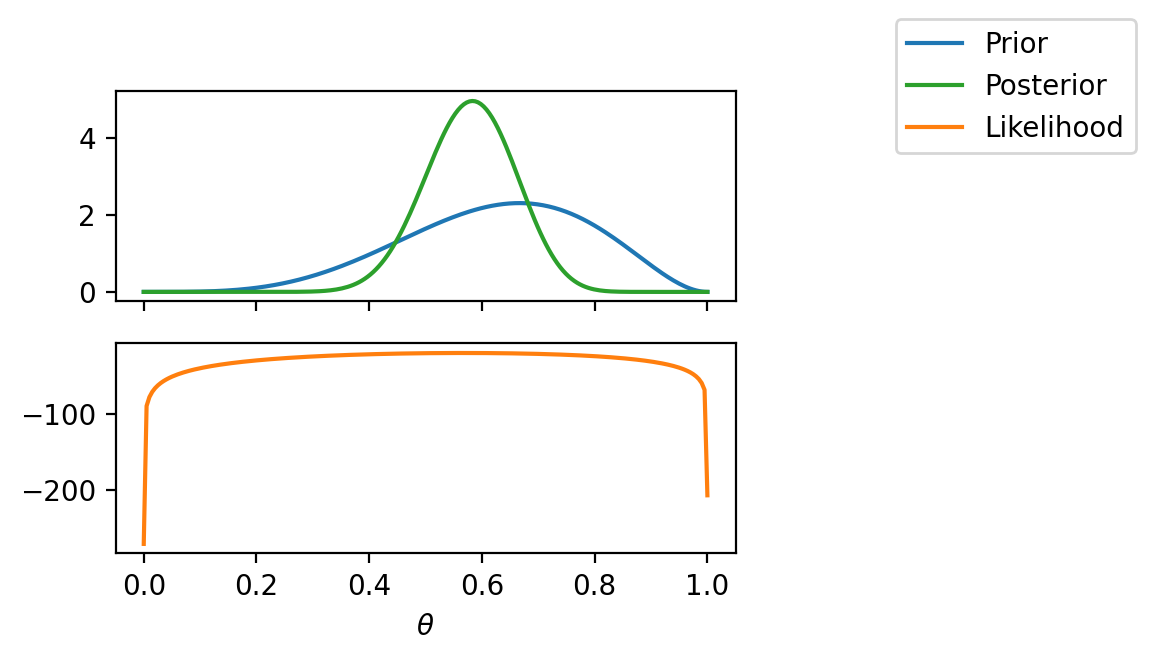
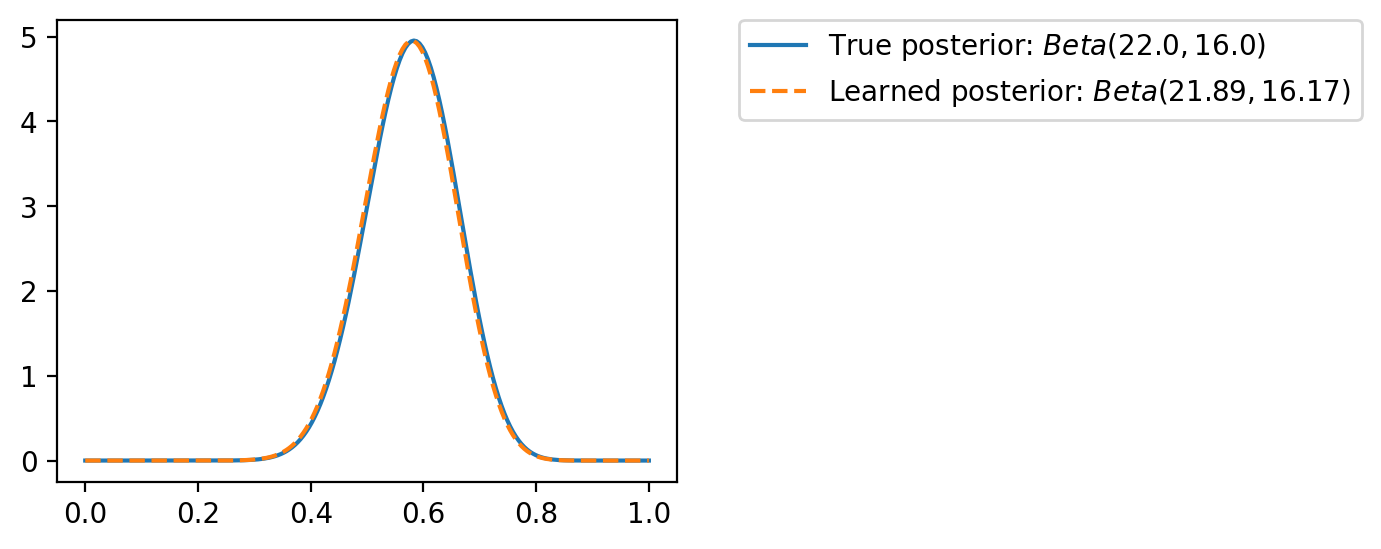
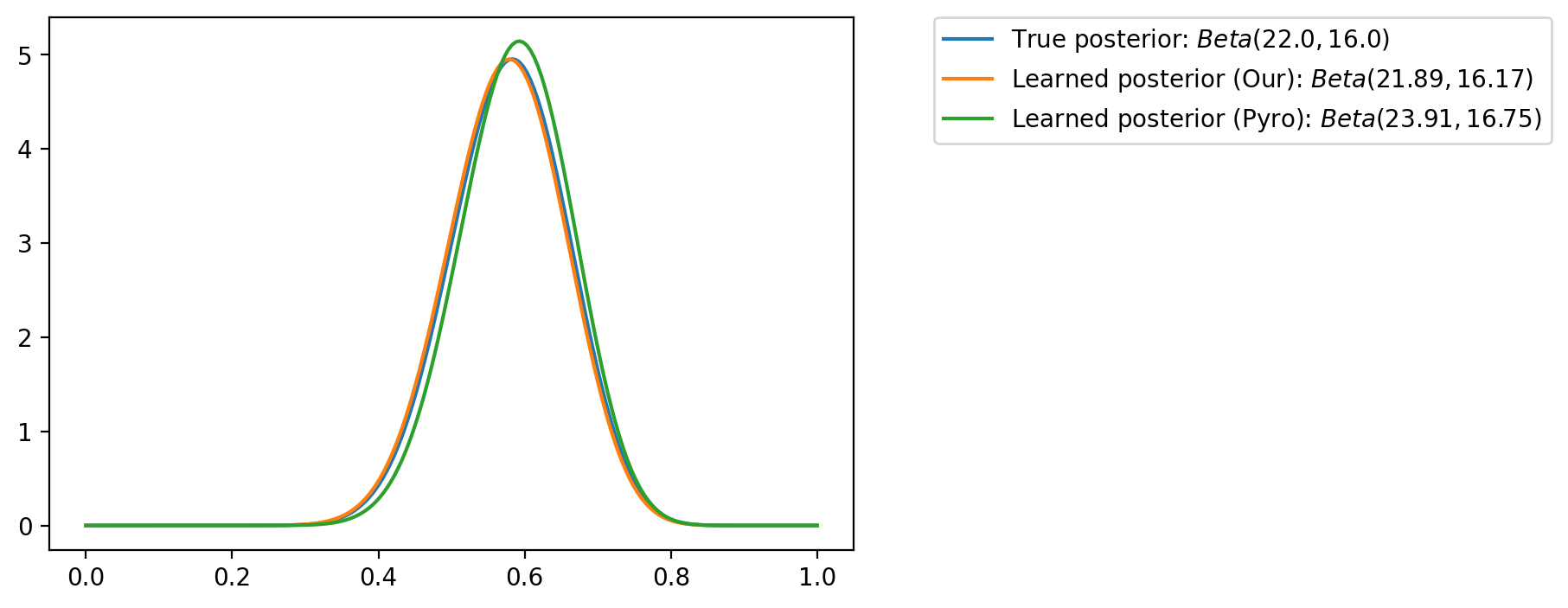
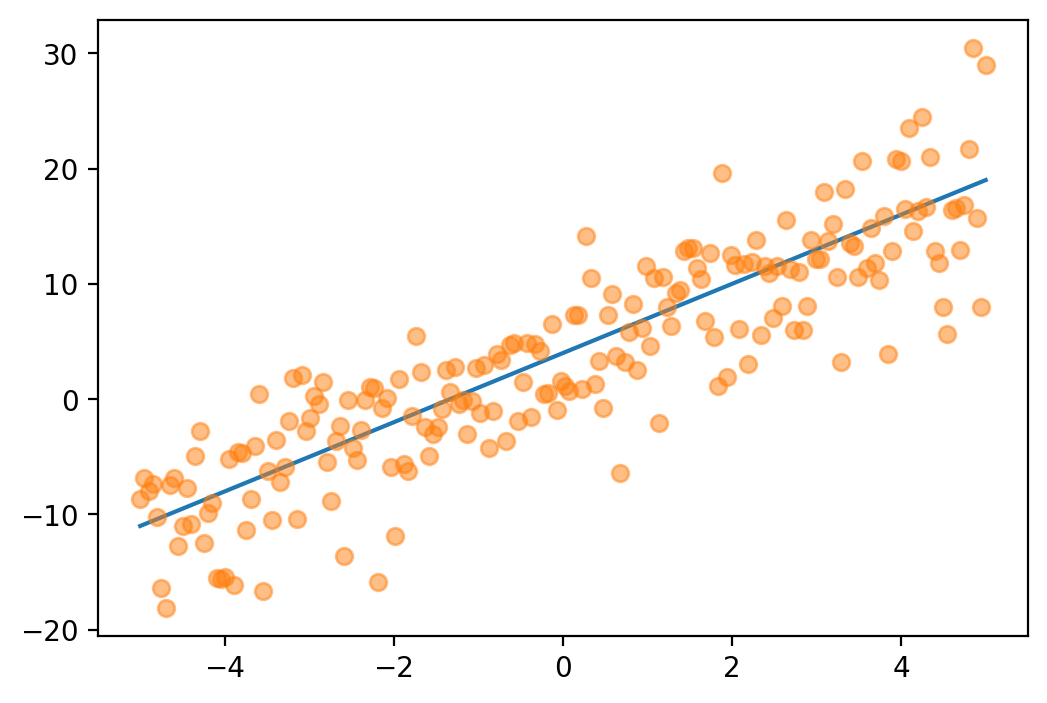
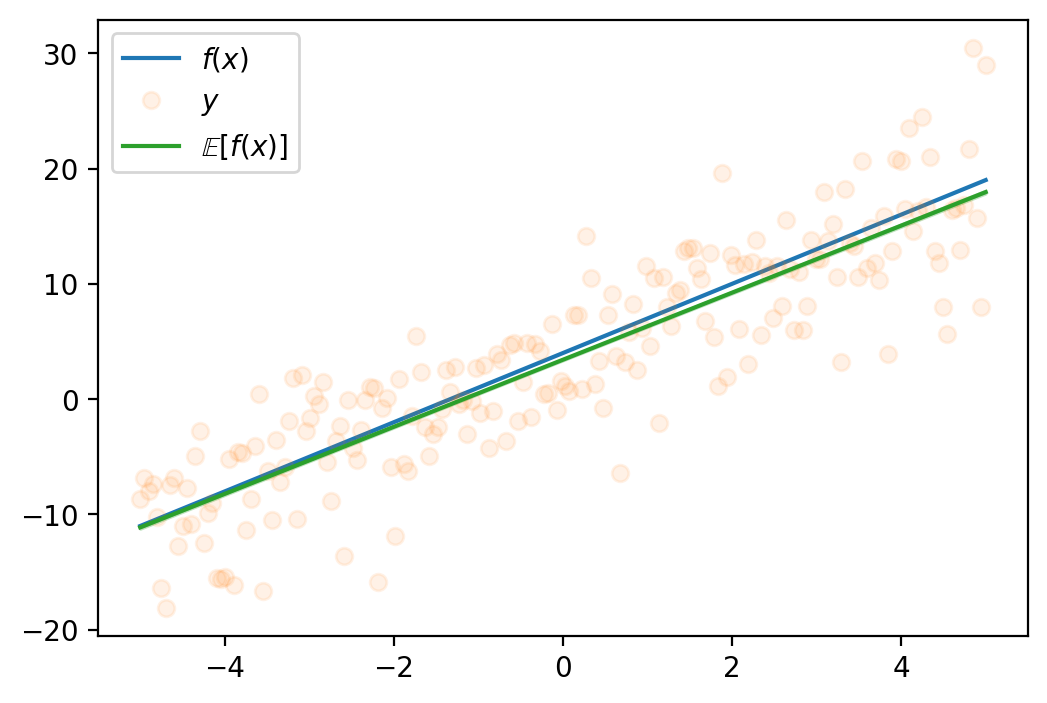
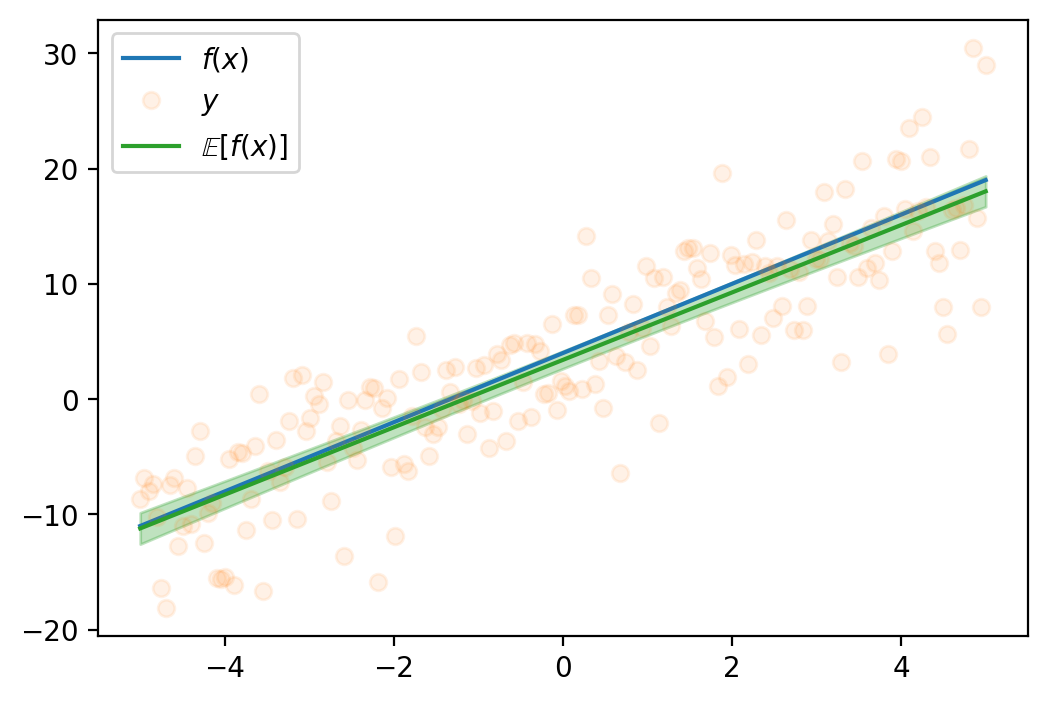
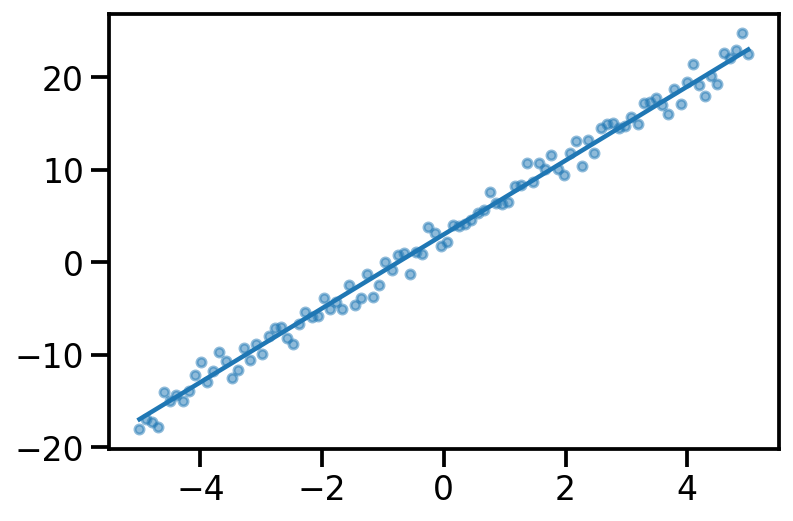
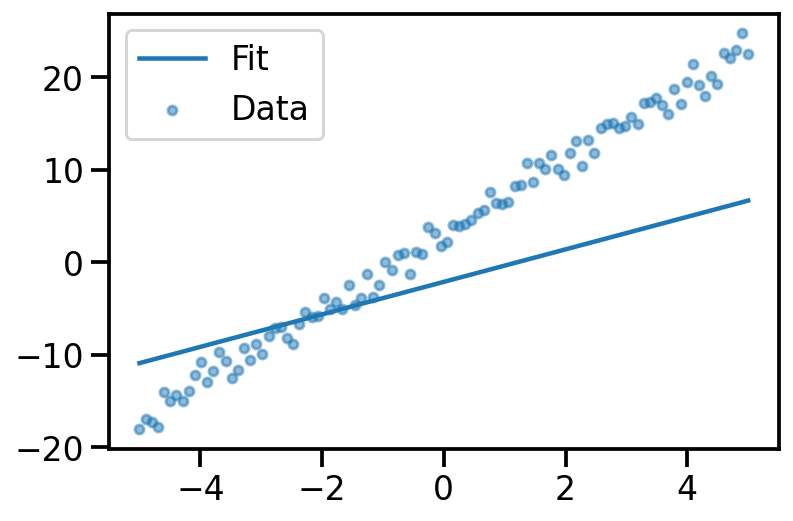
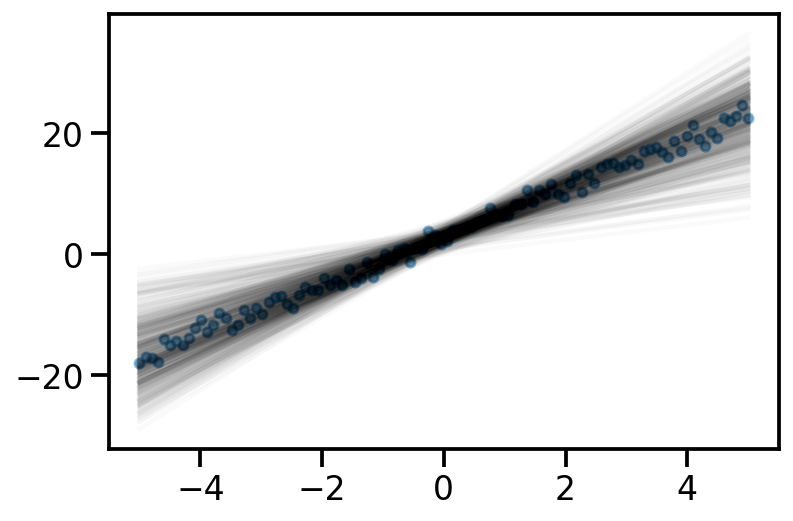
tensor([-1.6542e+01, -1.6148e+01, -1.5754e+01, -1.5360e+01, -1.4966e+01,
-1.4572e+01, -1.4178e+01, -1.3784e+01, -1.3390e+01, -1.2996e+01,
-1.2602e+01, -1.2208e+01, -1.1814e+01, -1.1420e+01, -1.1026e+01,
-1.0632e+01, -1.0238e+01, -9.8441e+00, -9.4501e+00, -9.0561e+00,
-8.6621e+00, -8.2681e+00, -7.8741e+00, -7.4801e+00, -7.0860e+00,
-6.6920e+00, -6.2980e+00, -5.9040e+00, -5.5100e+00, -5.1160e+00,
-4.7220e+00, -4.3280e+00, -3.9340e+00, -3.5400e+00, -3.1460e+00,
-2.7520e+00, -2.3579e+00, -1.9639e+00, -1.5699e+00, -1.1759e+00,
-7.8191e-01, -3.8790e-01, 6.1054e-03, 4.0011e-01, 7.9412e-01,
1.1881e+00, 1.5821e+00, 1.9761e+00, 2.3702e+00, 2.7642e+00,
3.1582e+00, 3.5522e+00, 3.9462e+00, 4.3402e+00, 4.7342e+00,
5.1282e+00, 5.5222e+00, 5.9162e+00, 6.3102e+00, 6.7043e+00,
7.0983e+00, 7.4923e+00, 7.8863e+00, 8.2803e+00, 8.6743e+00,
9.0683e+00, 9.4623e+00, 9.8563e+00, 1.0250e+01, 1.0644e+01,
1.1038e+01, 1.1432e+01, 1.1826e+01, 1.2220e+01, 1.2614e+01,
1.3008e+01, 1.3402e+01, 1.3796e+01, 1.4190e+01, 1.4584e+01,
1.4978e+01, 1.5372e+01, 1.5766e+01, 1.6160e+01, 1.6554e+01,
1.6948e+01, 1.7342e+01, 1.7736e+01, 1.8131e+01, 1.8525e+01,
1.8919e+01, 1.9313e+01, 1.9707e+01, 2.0101e+01, 2.0495e+01,
2.0889e+01, 2.1283e+01, 2.1677e+01, 2.2071e+01, 2.2465e+01])