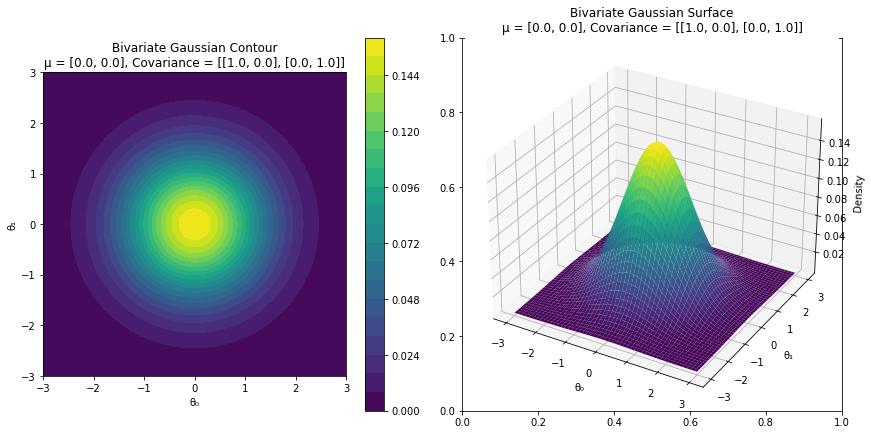
import torchimport matplotlib.pyplot as pltfrom matplotlib import cmfrom mpl_toolkits.mplot3d import Axes3Dimport numpy as npfrom ipywidgets import interact, FloatSliderdef visualize_bivariate_gaussian(mu_0, mu_1, cov_00, cov_01, cov_10, cov_11):= torch.tensor([mu_0, mu_1])= torch.tensor([[cov_00, cov_01],= torch.distributions.MultivariateNormal(mean, covariance_matrix)= 200 = torch.linspace(- 3 , 3 , N)= torch.linspace(- 3 , 3 , N)= torch.meshgrid(theta_0, theta_1)= torch.stack((Theta_0, Theta_1), dim= 2 )= torch.exp(bivariate_dist.log_prob(pos))= cm.get_cmap('viridis' )= plt.subplots(1 , 2 , figsize= (12 , 6 ))= axs[0 ].contourf(Theta_0, Theta_1, density, cmap= custom_cmap, levels= 20 )= axs[0 ])0 ].set_xlabel('θ \u2080 ' )0 ].set_ylabel('θ \u2081 ' )0 ].set_title(f'Bivariate Gaussian Contour \n μ = { mean. tolist()} , Covariance = { covariance_matrix. tolist()} ' )0 ].set_aspect('equal' ) # Set equal aspect ratio 1 ] = fig.add_subplot(122 , projection= '3d' )= axs[1 ].plot_surface(Theta_0, Theta_1, density, cmap= custom_cmap)1 ].set_xlabel('θ \u2080 ' )1 ].set_ylabel('θ \u2081 ' )1 ].set_zlabel('Density' )1 ].set_title(f'Bivariate Gaussian Surface \n μ = { mean. tolist()} , Covariance = { covariance_matrix. tolist()} ' )