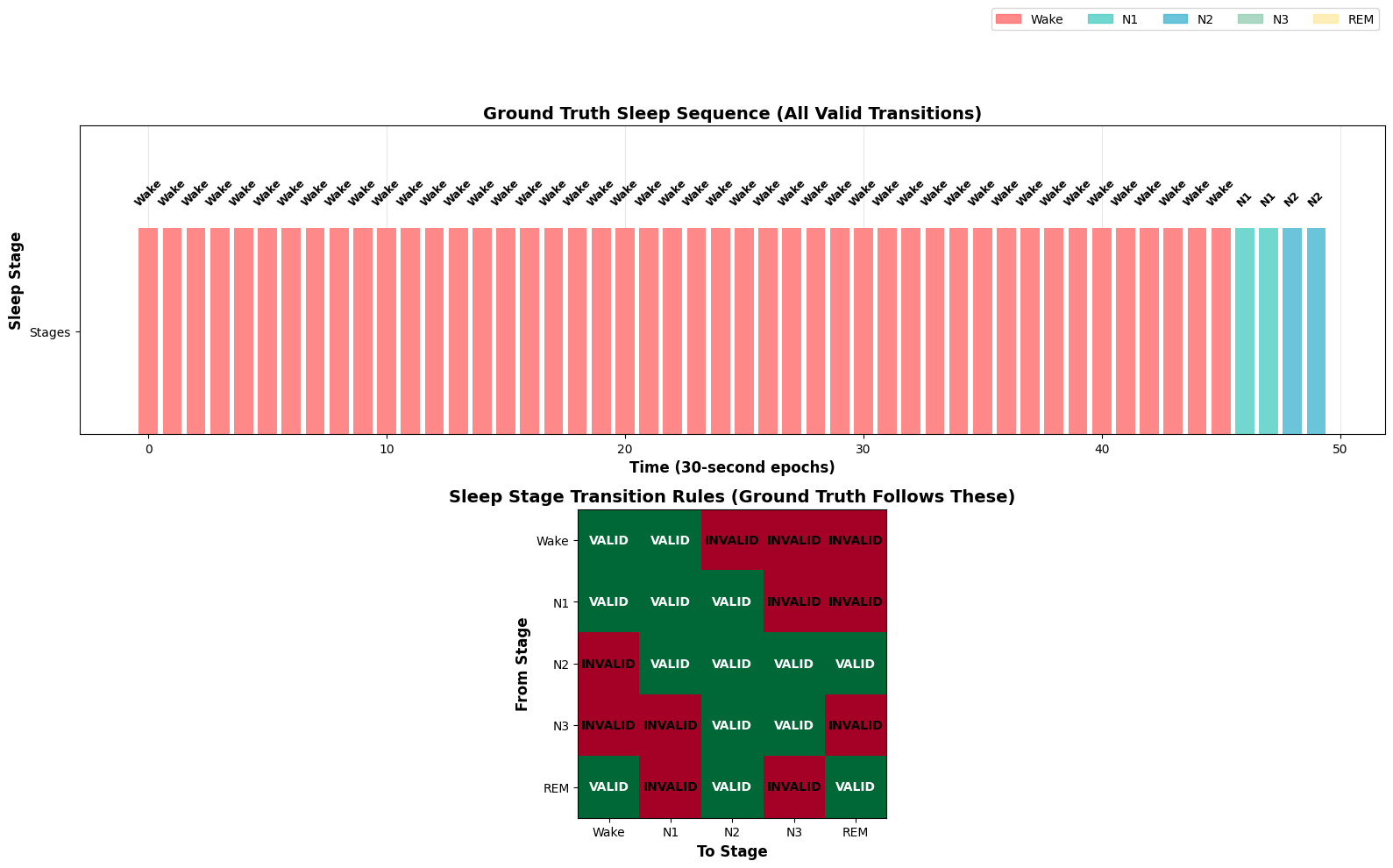
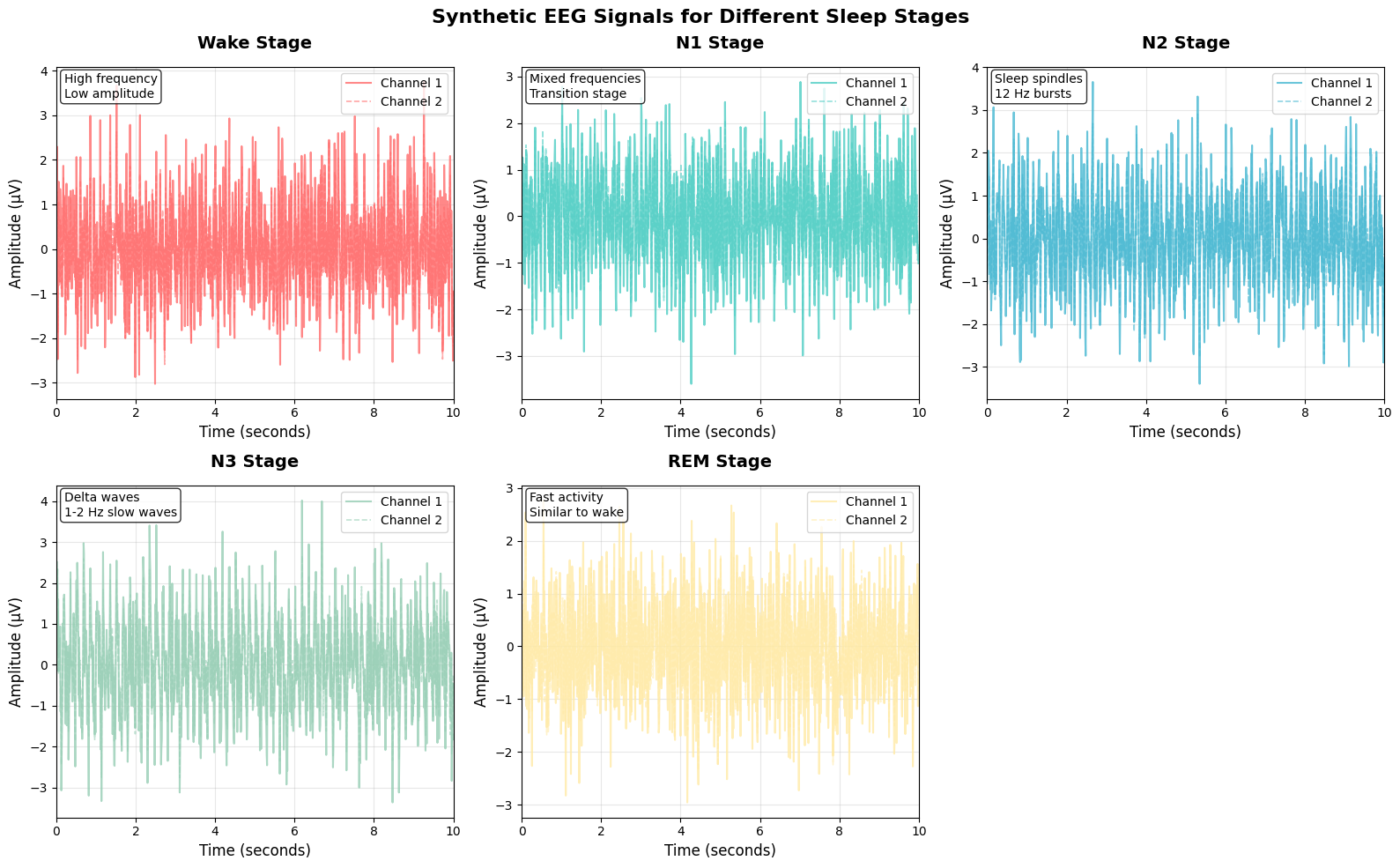
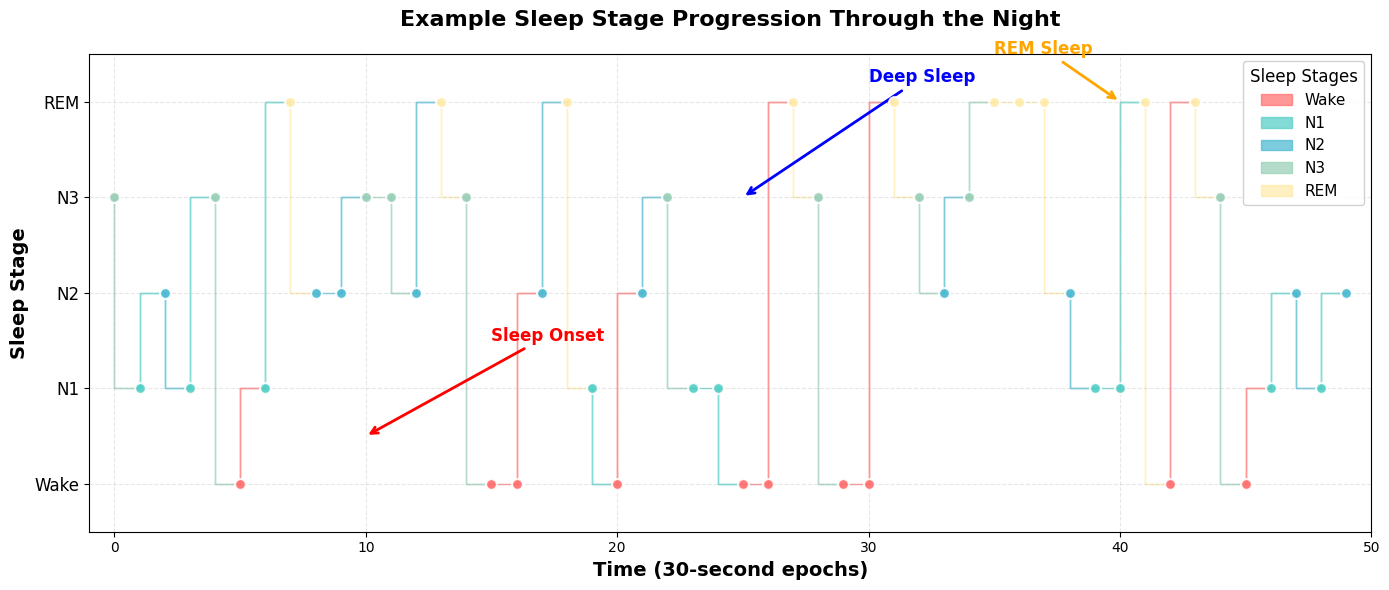
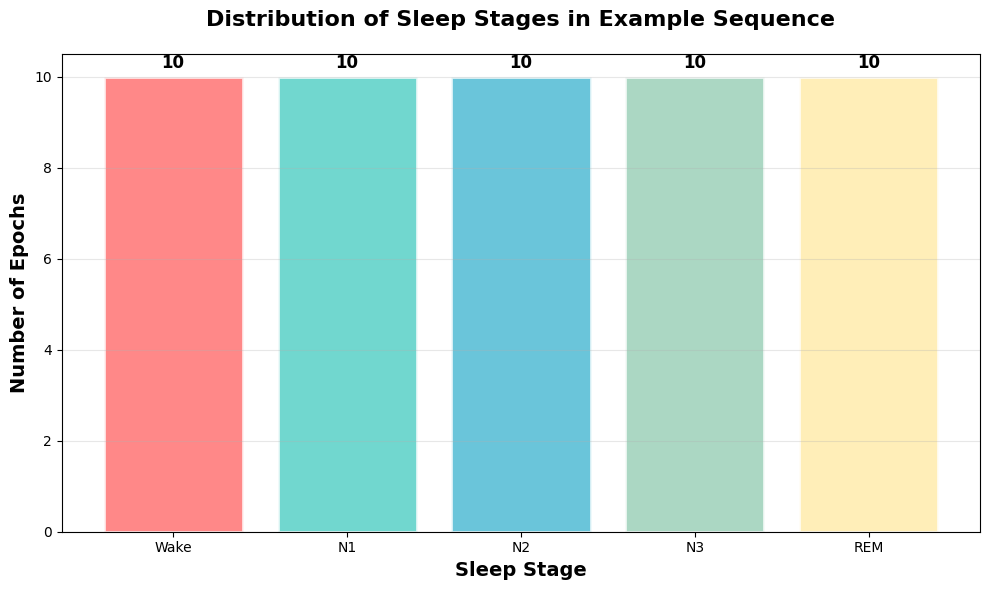
# Show sleep sequence example
example_seq = generate_sleep_sequence(50)
plt.figure(figsize=(14, 6))
plt.style.use('default') # Reset style
# Create a more informative plot
time_points = np.arange(len(example_seq))
stage_colors = ['#FF6B6B', '#4ECDC4', '#45B7D1', '#96CEB4', '#FFEAA7']
# Plot with filled areas for better visualization
for i in range(len(example_seq)-1):
plt.fill_between([i, i+1], [example_seq[i], example_seq[i+1]],
[example_seq[i], example_seq[i+1]],
color=stage_colors[example_seq[i]], alpha=0.7, step='pre')
# Add scatter points for clarity
for i, stage in enumerate(example_seq):
plt.scatter(i, stage, color=stage_colors[stage], s=60, alpha=0.9,
edgecolors='white', linewidth=1.5, zorder=5)
plt.xlabel('Time (30-second epochs)', fontsize=14, fontweight='bold')
plt.ylabel('Sleep Stage', fontsize=14, fontweight='bold')
plt.title('Example Sleep Stage Progression Through the Night', fontsize=16, fontweight='bold', pad=20)
# Customize y-axis
plt.yticks(range(5), STAGES, fontsize=12)
plt.grid(True, alpha=0.3, linestyle='--')
# Add annotations to explain the progression
plt.annotate('Sleep Onset', xy=(10, 0.5), xytext=(15, 1.5),
arrowprops=dict(arrowstyle='->', color='red', lw=2),
fontsize=12, fontweight='bold', color='red')
plt.annotate('Deep Sleep', xy=(25, 3), xytext=(30, 4.2),
arrowprops=dict(arrowstyle='->', color='blue', lw=2),
fontsize=12, fontweight='bold', color='blue')
plt.annotate('REM Sleep', xy=(40, 4), xytext=(35, 4.5),
arrowprops=dict(arrowstyle='->', color='orange', lw=2),
fontsize=12, fontweight='bold', color='orange')
# Add legend
legend_elements = [plt.Rectangle((0,0),1,1, color=stage_colors[i], alpha=0.7, label=stage)
for i, stage in enumerate(STAGES)]
plt.legend(handles=legend_elements, loc='upper right', fontsize=11,
title='Sleep Stages', title_fontsize=12, framealpha=0.9)
plt.xlim(-1, len(example_seq))
plt.ylim(-0.5, 4.5)
plt.tight_layout()
plt.show()
# Show stage distribution
stage_counts = [example_seq.count(i) for i in range(5)]
plt.figure(figsize=(10, 6))
bars = plt.bar(STAGES, stage_counts, color=stage_colors, alpha=0.8, edgecolor='white', linewidth=2)
plt.xlabel('Sleep Stage', fontsize=14, fontweight='bold')
plt.ylabel('Number of Epochs', fontsize=14, fontweight='bold')
plt.title('Distribution of Sleep Stages in Example Sequence', fontsize=16, fontweight='bold', pad=20)
plt.grid(True, alpha=0.3, axis='y')
# Add value labels on bars
for bar, count in zip(bars, stage_counts):
height = bar.get_height()
plt.text(bar.get_x() + bar.get_width()/2., height + 0.1,
f'{count}', ha='center', va='bottom', fontsize=12, fontweight='bold')
plt.tight_layout()
plt.show()