import numpy as np
import matplotlib.pyplot as plt
import torch
import seaborn as sns
import pandas as pd
dist =torch.distributions
sns.reset_defaults()
sns.set_context(context="talk", font_scale=1)
%matplotlib inline
%config InlineBackend.figure_format='retina'Basic Imports
# Generate complex data
x = np.linspace(-2, 2, 100)
freq = lambda x: x**2
f = lambda x: np.sin(2 * np.pi * x * freq(x))
y = f(x) + np.random.randn(x.shape[0]) * 0.1
# Plot
plt.figure(figsize=(8, 5))
plt.plot(x, y, 'o', label='data')
plt.plot(x, f(x), label='true function')
plt.legend()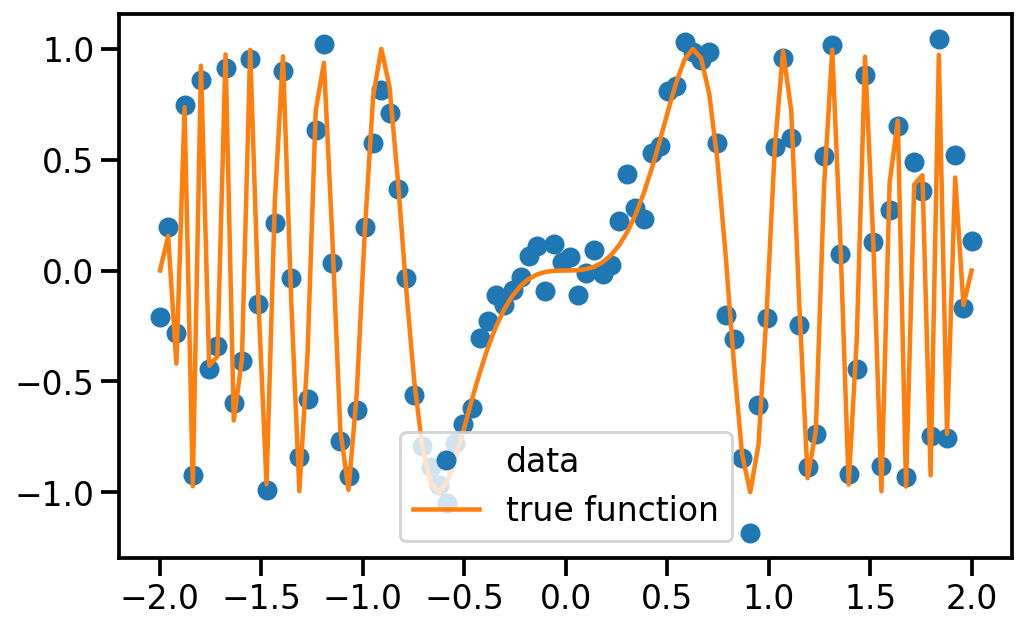
# Learn linear model on data
from sklearn.linear_model import LinearRegression, Ridge
lr1 = Ridge()
lr1.fit(x.reshape(-1, 1), y.reshape(-1, 1))
# Predict on linspace and plot
y_pred = lr1.predict(x.reshape(-1, 1))
plt.figure(figsize=(8, 5))
plt.plot(x, y, 'o', label='data')
plt.plot(x, f(x), label='true function')
plt.plot(x, y_pred, label='linear model')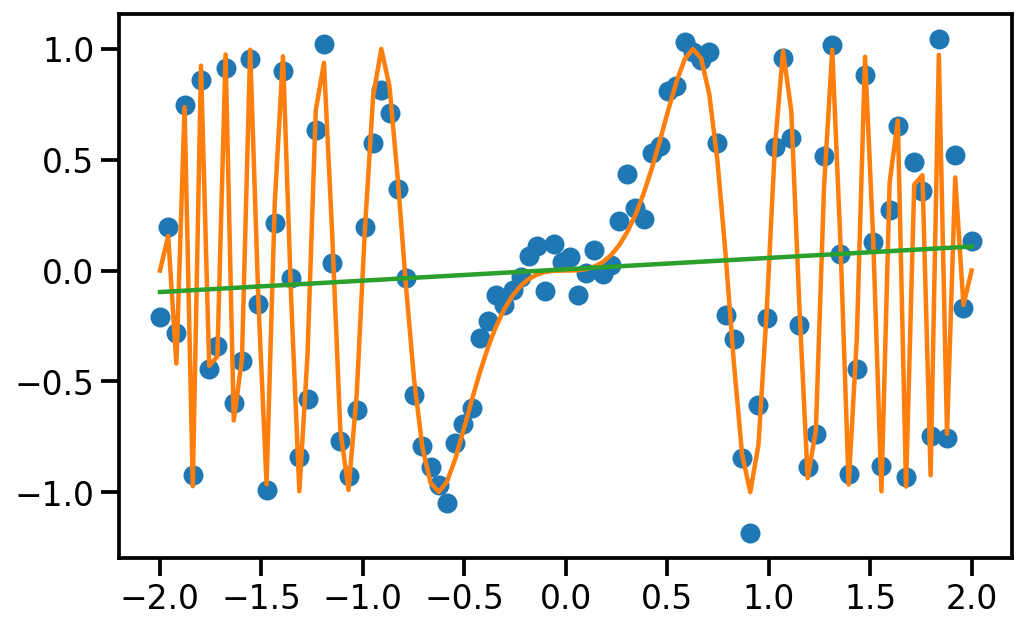
# Add position encoding
# Gamma(x) = [sin(2^0*pi*x), cos(2^0*pi*x), sin(2^1*pi*x), cos(2^1*pi*x), ..., sin(2^k*pi*x), cos(2^k*pi*x)]
def gamma(x, k):
"""
x: (N, 1)
k: int
Output: (N, 2k)
"""
x = x.reshape(-1, 1)
x = np.repeat(x, k, axis=1)
x = x * (2 ** np.arange(k) * np.pi)
z = np.concatenate([np.sin(x), np.cos(x)], axis=1)
# Concatenate x and z
z = np.concatenate([x, z], axis=1)
return z
plt.plot(x, gamma(x, 2), alpha=0.2, color='k')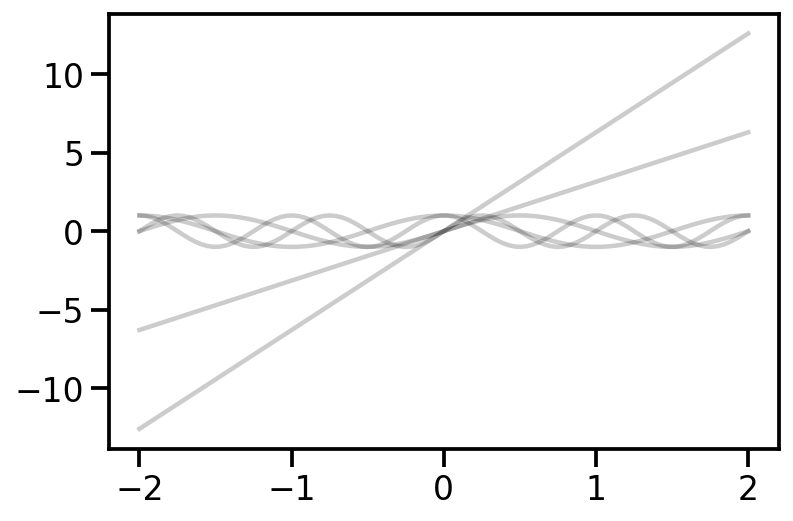
# Fit linear model with position encoding for k
def fit_plot(x, k):
lr = Ridge()
X_new = gamma(x, k)
lr.fit(X_new, y.reshape(-1, 1))
y_pred = lr.predict(X_new)
plt.figure(figsize=(8, 5))
plt.plot(x, y, 'o', label='data')
plt.plot(x, f(x), label='true function')
plt.plot(x, y_pred, label='linear model with position encoding of order {}'.format(k), lw=3)
# Legend outside plot
plt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)
# Title is the score
plt.title('Score: {:.2f}'.format(lr.score(X_new, y.reshape(-1, 1))))
# Show extrapolation also
# extrapolation points are set difference between linspace and data points
x_extra = np.linspace(-3, 3, 100)
X_extra = gamma(x_extra, k)
y_extra = lr.predict(X_extra)
plt.plot(x_extra, y_extra, '--', label='extrapolation', alpha=0.2)
plt.legend(bbox_to_anchor=(1.05, 1), loc=2, borderaxespad=0.)
fit_plot(x, 1)--------------------------------------------------------------------------- ValueError Traceback (most recent call last) /home/nipun.batra/git/blog/posts/positional-encoding.ipynb Cell 8 in <module> ----> <a href='vscode-notebook-cell://ssh-remote%2B10.0.62.168/home/nipun.batra/git/blog/posts/positional-encoding.ipynb#X36sdnNjb2RlLXJlbW90ZQ%3D%3D?line=0'>1</a> fit_plot(x, 1) /home/nipun.batra/git/blog/posts/positional-encoding.ipynb Cell 8 in fit_plot(x, k) <a href='vscode-notebook-cell://ssh-remote%2B10.0.62.168/home/nipun.batra/git/blog/posts/positional-encoding.ipynb#X36sdnNjb2RlLXJlbW90ZQ%3D%3D?line=3'>4</a> lr = Ridge() <a href='vscode-notebook-cell://ssh-remote%2B10.0.62.168/home/nipun.batra/git/blog/posts/positional-encoding.ipynb#X36sdnNjb2RlLXJlbW90ZQ%3D%3D?line=4'>5</a> X_new = gamma(x, k) ----> <a href='vscode-notebook-cell://ssh-remote%2B10.0.62.168/home/nipun.batra/git/blog/posts/positional-encoding.ipynb#X36sdnNjb2RlLXJlbW90ZQ%3D%3D?line=5'>6</a> lr.fit(X_new, y.reshape(-1, 1)) <a href='vscode-notebook-cell://ssh-remote%2B10.0.62.168/home/nipun.batra/git/blog/posts/positional-encoding.ipynb#X36sdnNjb2RlLXJlbW90ZQ%3D%3D?line=6'>7</a> y_pred = lr.predict(X_new) <a href='vscode-notebook-cell://ssh-remote%2B10.0.62.168/home/nipun.batra/git/blog/posts/positional-encoding.ipynb#X36sdnNjb2RlLXJlbW90ZQ%3D%3D?line=7'>8</a> plt.figure(figsize=(8, 5)) File ~/miniforge3/lib/python3.9/site-packages/sklearn/linear_model/_ridge.py:1003, in Ridge.fit(self, X, y, sample_weight) 983 """Fit Ridge regression model. 984 985 Parameters (...) 1000 Fitted estimator. 1001 """ 1002 _accept_sparse = _get_valid_accept_sparse(sparse.issparse(X), self.solver) -> 1003 X, y = self._validate_data( 1004 X, 1005 y, 1006 accept_sparse=_accept_sparse, 1007 dtype=[np.float64, np.float32], 1008 multi_output=True, 1009 y_numeric=True, 1010 ) 1011 return super().fit(X, y, sample_weight=sample_weight) File ~/miniforge3/lib/python3.9/site-packages/sklearn/base.py:581, in BaseEstimator._validate_data(self, X, y, reset, validate_separately, **check_params) 579 y = check_array(y, **check_y_params) 580 else: --> 581 X, y = check_X_y(X, y, **check_params) 582 out = X, y 584 if not no_val_X and check_params.get("ensure_2d", True): File ~/miniforge3/lib/python3.9/site-packages/sklearn/utils/validation.py:981, in check_X_y(X, y, accept_sparse, accept_large_sparse, dtype, order, copy, force_all_finite, ensure_2d, allow_nd, multi_output, ensure_min_samples, ensure_min_features, y_numeric, estimator) 964 X = check_array( 965 X, 966 accept_sparse=accept_sparse, (...) 976 estimator=estimator, 977 ) 979 y = _check_y(y, multi_output=multi_output, y_numeric=y_numeric) --> 981 check_consistent_length(X, y) 983 return X, y File ~/miniforge3/lib/python3.9/site-packages/sklearn/utils/validation.py:332, in check_consistent_length(*arrays) 330 uniques = np.unique(lengths) 331 if len(uniques) > 1: --> 332 raise ValueError( 333 "Found input variables with inconsistent numbers of samples: %r" 334 % [int(l) for l in lengths] 335 ) ValueError: Found input variables with inconsistent numbers of samples: [100, 768]
fit_plot(x, 2)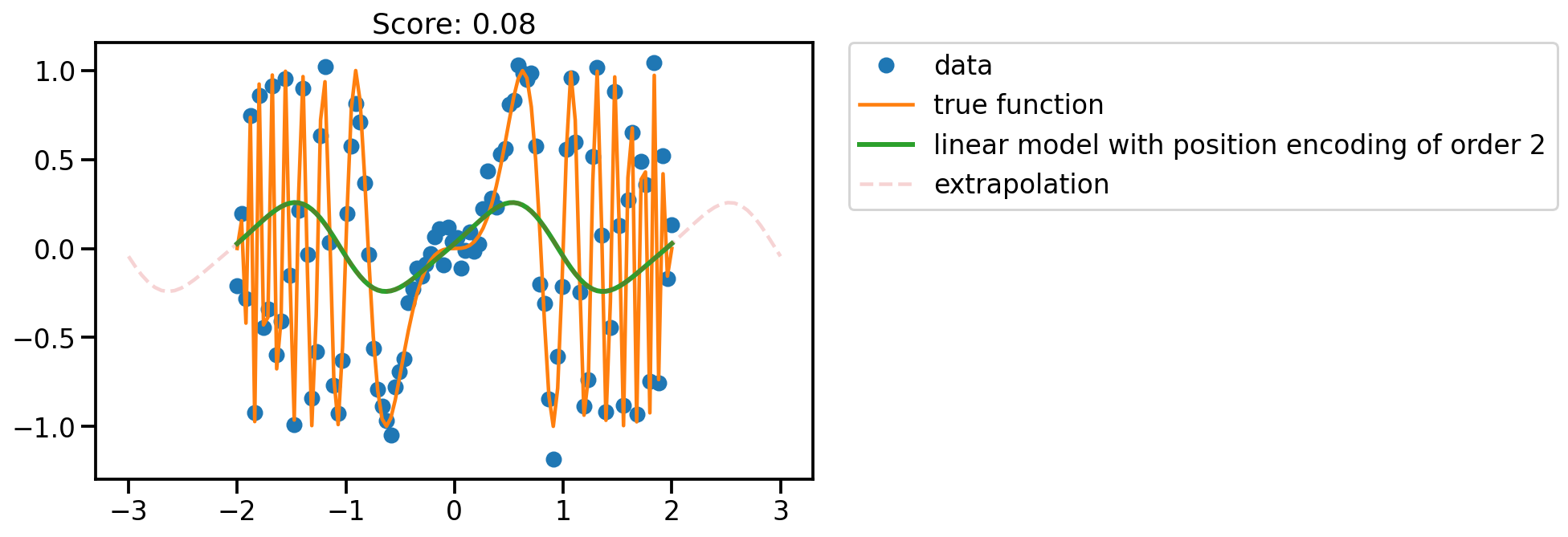
fit_plot(x, 3)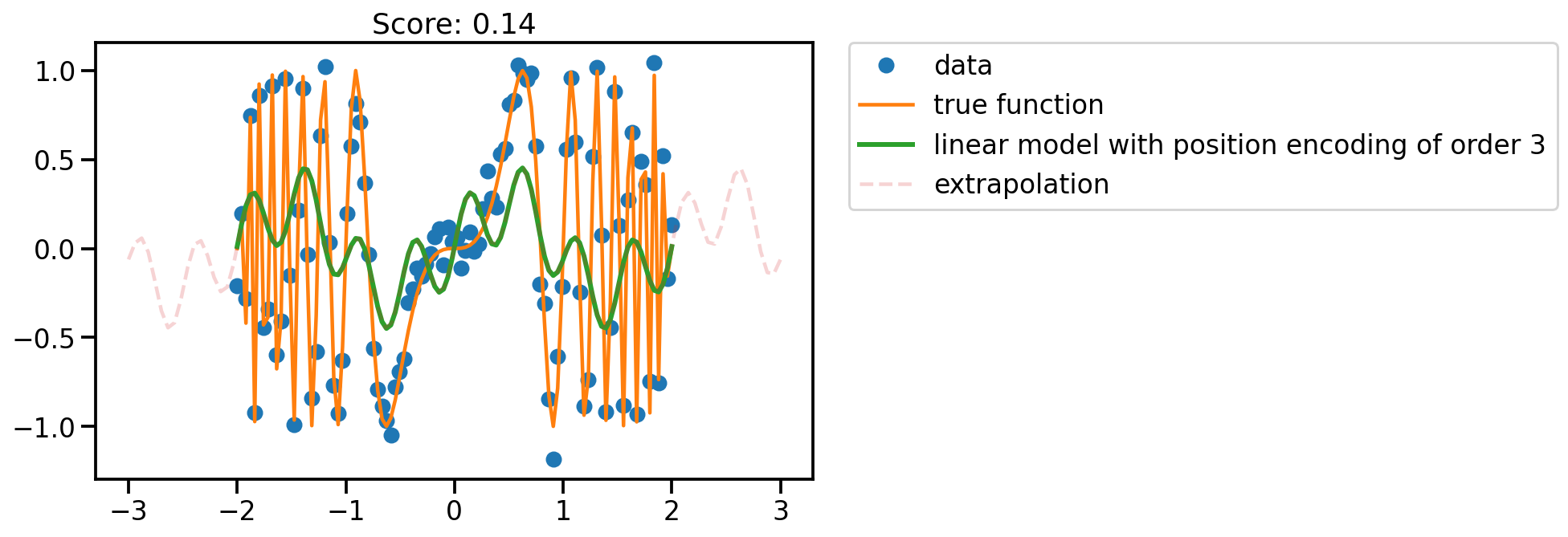
fit_plot(x, 7)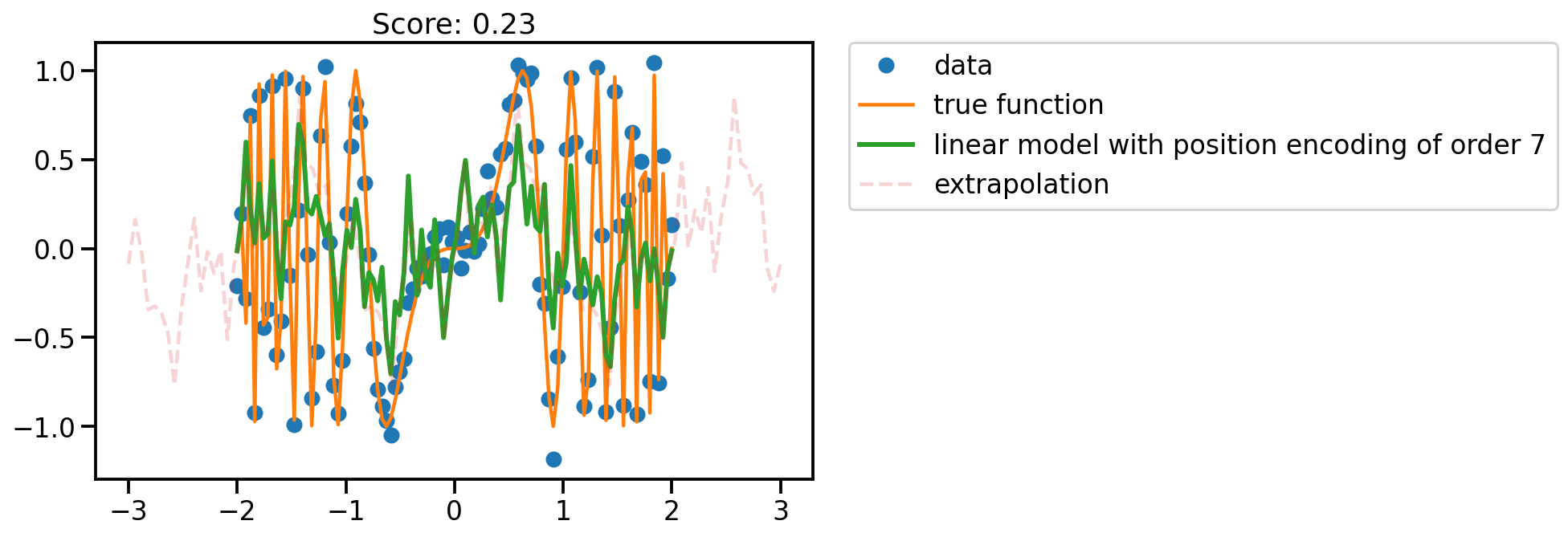
fit_plot(x, 10)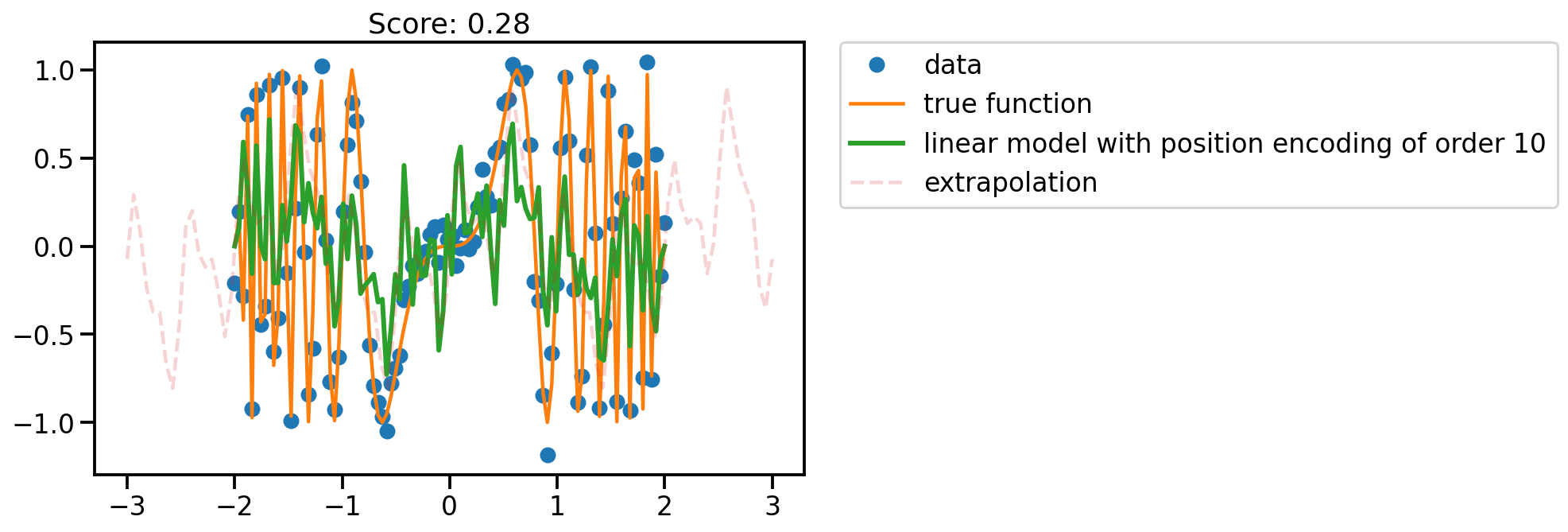
fit_plot(x, 70)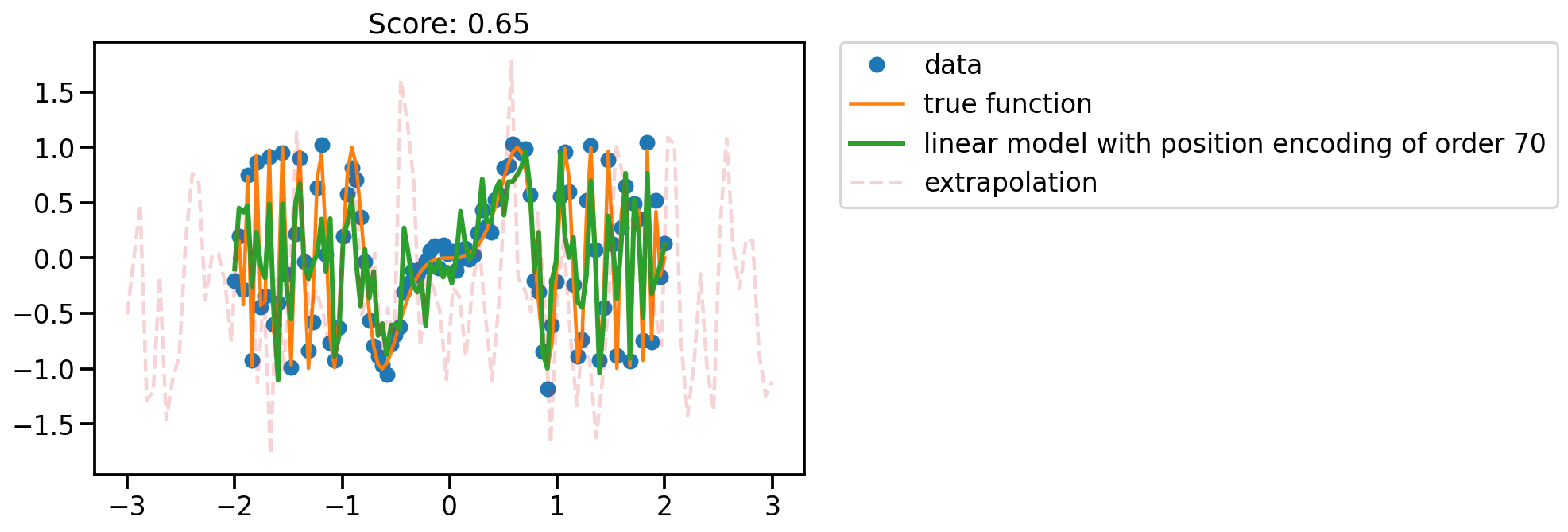
!wget https://gml.noaa.gov/webdata/ccgg/trends/co2/co2_mm_mlo.csv--2023-06-09 15:39:49-- https://gml.noaa.gov/webdata/ccgg/trends/co2/co2_mm_mlo.csv
Resolving gml.noaa.gov (gml.noaa.gov)... 140.172.200.41, 2610:20:8800:6101::29
Connecting to gml.noaa.gov (gml.noaa.gov)|140.172.200.41|:443... connected.
HTTP request sent, awaiting response... 200 OK
Length: 38018 (37K) [text/csv]
Saving to: ‘co2_mm_mlo.csv’
co2_mm_mlo.csv 100%[===================>] 37.13K 146KB/s in 0.3s
2023-06-09 15:39:51 (146 KB/s) - ‘co2_mm_mlo.csv’ saved [38018/38018]
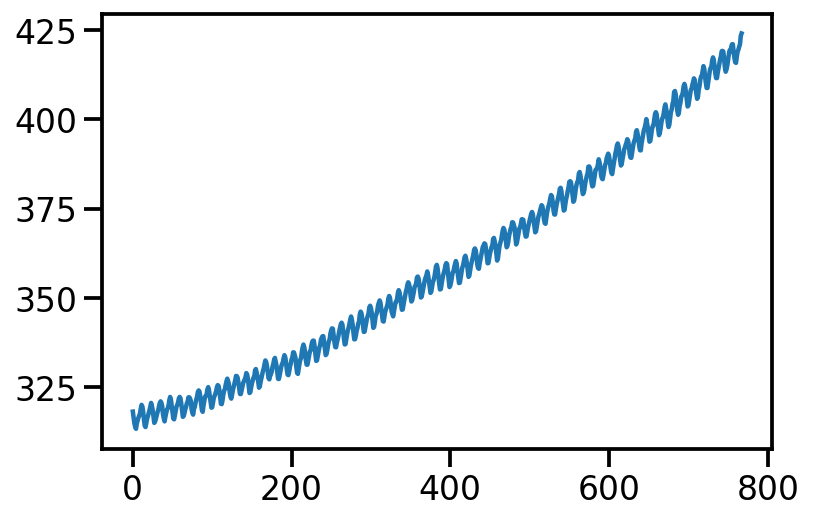
df = pd.read_csv('co2_mm_mlo.csv', header=None, skiprows=72)X = df.index.values
y = df[3].values.reshape(-1, 1)plt.plot(X, y)