Probability Mass Functions and Common Discrete Distributions
probability mass function, PMF, discrete distributions, bernoulli distribution, categorical distribution, binomial distribution, poisson distribution, geometric distribution, quality control, logistic regression
Probability Mass Functions and Common Discrete Distributions
Learning Objectives
By the end of this notebook, you will understand:
- Probability Mass Functions (PMF): Mathematical definition and properties
- Common Discrete Distributions: Bernoulli, Categorical, Binomial, Poisson, and Geometric
- Real-World Applications: Quality control, classification, and count modeling
- Parameter Estimation: Fitting distributions to data using PyTorch
- Model Selection: Choosing appropriate distributions for different scenarios
- Practical Implementation: Working with PyTorch distributions for data science
Introduction
Discrete random variables take on countable values (like integers), and their behavior is completely described by their Probability Mass Function (PMF). The PMF tells us the probability of each possible outcome, providing a complete probabilistic picture of the random variable.
Mathematical Foundation
For a discrete random variable \(X\), the PMF is defined as:
\[p_X(x) = P(X = x)\]
The PMF must satisfy two fundamental properties: 1. Non-negativity: \(p_X(x) \geq 0\) for all \(x\) 2. Normalization: \(\sum_{x} p_X(x) = 1\)
Why Discrete Distributions Matter
Discrete distributions are everywhere in data science and statistics: - Classification problems (Bernoulli for binary, Categorical for multi-class) - Count data (Poisson for rare events, Binomial for fixed trials) - Quality control (Binomial for defect rates) - Waiting times (Geometric for time to first success) - Natural language processing (Categorical for word distributions)
Common Discrete Distributions Overview
We’ll explore these essential distributions:
| Distribution | Use Case | Parameters | Example |
|---|---|---|---|
| Bernoulli | Binary outcomes | \(p\) (success prob) | Coin flip, Pass/Fail |
| Categorical | Multi-class outcomes | \(\boldsymbol{\theta}\) (prob vector) | Die roll, Image classification |
| Binomial | Fixed trials, binary outcome | \(n, p\) | Quality control, Survey responses |
| Poisson | Count of rare events | \(\lambda\) (rate) | Email arrivals, Defects per unit |
| Geometric | Trials to first success | \(p\) (success prob) | Sales calls, Equipment failure |
Let’s dive into each distribution with theory, implementation, and real-world applications!
Setting Up the Environment
We’ll use PyTorch for probability distributions, NumPy for numerical operations, and Matplotlib for visualizations:
1. Bernoulli Distribution
The Bernoulli distribution is the simplest discrete distribution, modeling a single binary trial with two possible outcomes: success (1) or failure (0).
Mathematical Definition
Let \(X\) be a Bernoulli random variable with parameter \(p \in [0,1]\). The PMF is:
\[p_X(x) = \begin{cases} 1-p, & \text{if } x = 0 \\ p, & \text{if } x = 1 \end{cases}\]
Equivalently: \(p_X(x) = p^x(1-p)^{1-x}\) for \(x \in \{0, 1\}\)
Properties
- Mean: \(E[X] = p\)
- Variance: \(\text{Var}(X) = p(1-p)\)
- Maximum variance: Occurs when \(p = 0.5\)
Visualization
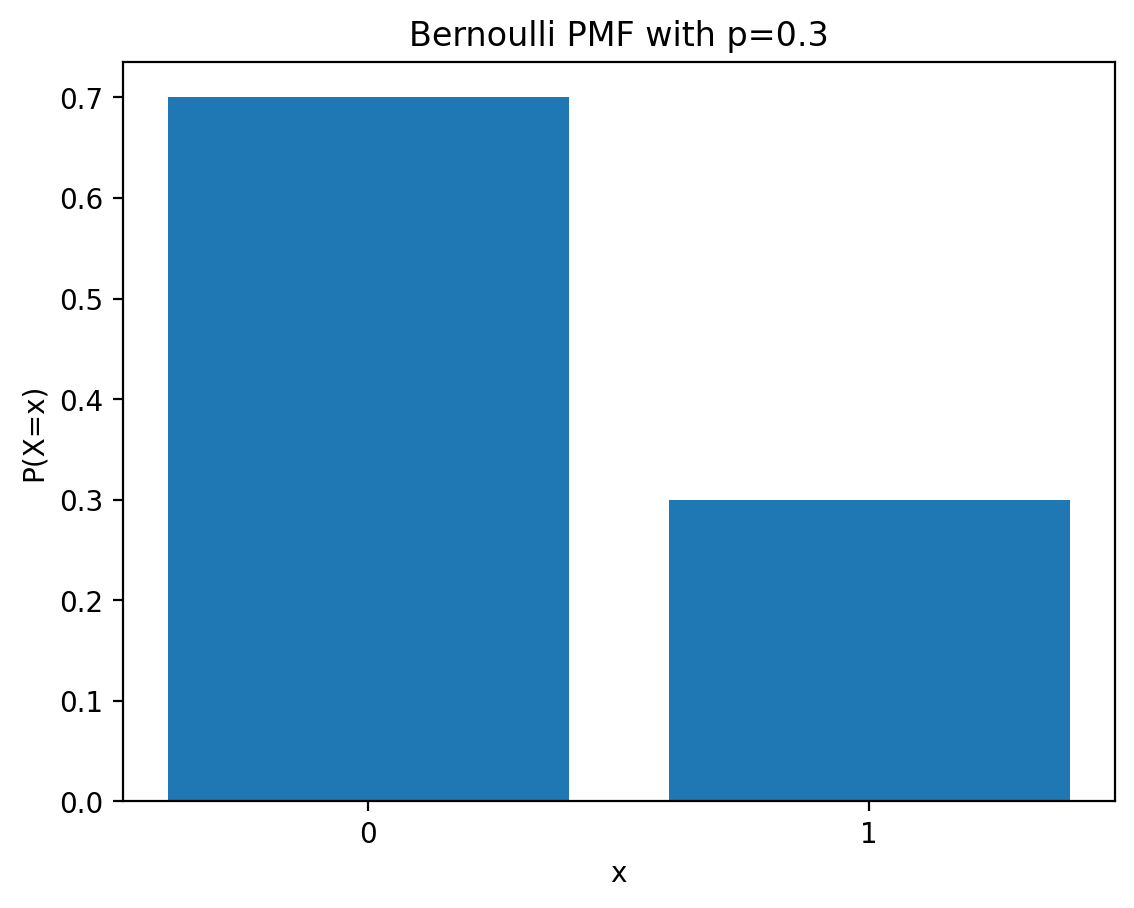
PMF of Bernoulli distribution
Let \(X\) be a Bernoulli random variable. \(X\) can take on one of two values, 0 or 1, with probabilities \(1-p\) and \(p\), respectively.
Example: Suppose we flip a coin with probability \(p\) of landing heads. Let \(X\) be the random variable that is 1 if the coin lands heads and 0 if the coin lands tails.
The probability mass function (PMF) of a Bernoulli random variable is given by:
\[ p_X(x) = \begin{cases} 1-p, & \text{if } x = 0, \\ p, & \text{if } x = 1. \end{cases} \]
or, equivalently,
\[ p_X(x) = p^x(1-p)^{1-x}, \quad x \in \{0, 1\}. \]
where \(0 < p < 1\) is called the Bernoulli parameter. We write
\[ X \sim \text{Bernoulli}(p) \]
to denote that \(X\) is drawn from a Bernoulli distribution with parameter \(p\).
The probability mass function (PMF) of a Bernoulli distribution is given by: \[ \begin{equation} f(x) = \begin{cases} p & \text{if } x = 1 \\ 1 - p & \text{if } x = 0 \end{cases} \end{equation} \]
where \(p\) is the probability of success.
Working with PyTorch Distributions
Let’s explore the Bernoulli distribution using PyTorch’s built-in functionality:
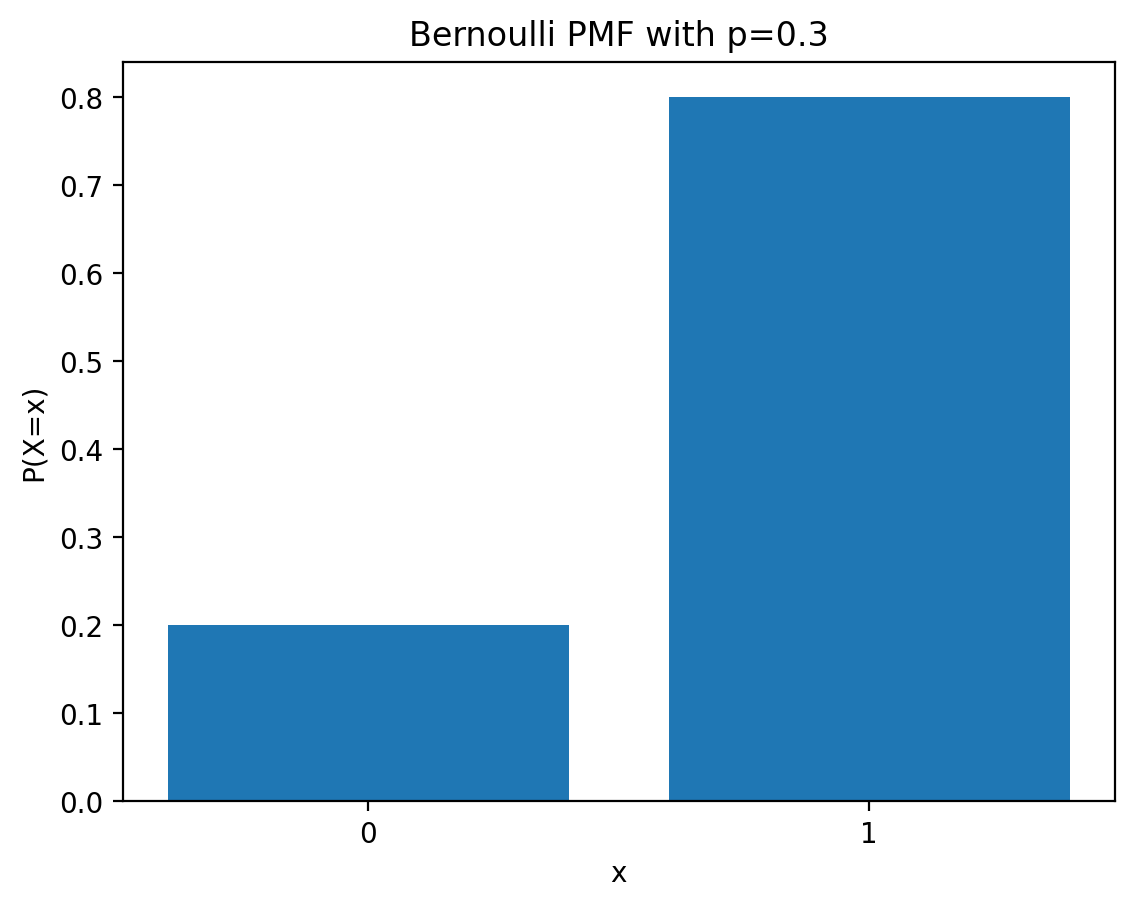
Init signature: torch.distributions.Bernoulli(probs=None, logits=None, validate_args=None) Docstring: Creates a Bernoulli distribution parameterized by :attr:`probs` or :attr:`logits` (but not both). Samples are binary (0 or 1). They take the value `1` with probability `p` and `0` with probability `1 - p`. Example:: >>> # xdoctest: +IGNORE_WANT("non-deterministic") >>> m = Bernoulli(torch.tensor([0.3])) >>> m.sample() # 30% chance 1; 70% chance 0 tensor([ 0.]) Args: probs (Number, Tensor): the probability of sampling `1` logits (Number, Tensor): the log-odds of sampling `1` File: ~/mambaforge/lib/python3.12/site-packages/torch/distributions/bernoulli.py Type: type Subclasses:
Bernoulli Distribution in PyTorch
# Print all attributes of the Bernoulli distribution -- do not have __ or _ in the beginning
attrs = [attr for attr in dir(dist) if not attr.startswith('_')]
pd.Series(attrs)0 arg_constraints
1 batch_shape
2 cdf
3 entropy
4 enumerate_support
5 event_shape
6 expand
7 has_enumerate_support
8 has_rsample
9 icdf
10 log_prob
11 logits
12 mean
13 mode
14 param_shape
15 perplexity
16 probs
17 rsample
18 sample
19 sample_n
20 set_default_validate_args
21 stddev
22 support
23 variance
dtype: objectExpected value argument (Tensor of shape ()) to be within the support (Boolean()) of the distribution Bernoulli(probs: 0.8999999761581421, logits: 2.1972243785858154), but found invalid values:
0.5Notice that even though the expected frequency is 0.9, individual samples can vary significantly due to randomness.
Real-World Application: Fruit Classification
Let’s explore a practical machine learning application using the Bernoulli distribution for binary classification.
Problem Setup: Lime vs Lemon Classification
We want to classify fruits as limes (0) or lemons (1) based on their radius. This is a classic logistic regression problem where we model:
\[P(\text{Lemon}|\text{radius}) = \text{sigmoid}(w \cdot \text{radius} + b)\]
The sigmoid function ensures probabilities stay between 0 and 1, making it perfect for Bernoulli distributions.
Lime vs Lemon

https://www.healthline.com/nutrition/lime-vs-lemon
Limes are small, green, and more tart than lemons, which are larger, oval-shaped, and yellow. Nutritionally, they’re almost identical and share many of the same potential health benefits.
Lemons are usually bright yellow, while limes are typically a bright shade of green. However, certain types of limes will turn yellow as they ripen, making the distinction a little more difficult.
Limes are also smaller and rounder than lemons. They can vary in size but are usually 1–2 inches (3–6 centimeters) in diameter.
In comparison, lemons tend to be 2–4 inches (7–12 centimeters) in diameter and have a more oval or oblong shape.
Main question
Given a fruit (lime or lemon) and its radius, we want to predict if it is a lime or a lemon.
Let us denote the radius of the fruit by \(r\) and the type of the fruit by \(y\) where \(y=0\) if the fruit is a lime and \(y=1\) if the fruit is a lemon.
We want to model the probability of the fruit being a lemon given its radius, i.e., we want to model \(p(y=1|r)\).
Generative process
tensor([2.6468, 2.7450, 1.1486, 2.8779, 1.1713, 1.8027, 0.7697, 2.3809, 2.8223,
0.3996])We start by modeling the generative process of the data.
We assume if w*r + b > 0, then the fruit is a lemon, otherwise it is a lime.
Let us assume that w_true = 1.2 and b_true = -2.0.
count 1000.000000
mean -0.249277
std 1.045096
min -1.991556
25% -1.134019
50% -0.342291
75% 0.646800
max 1.599296
dtype: float64Can we use logits to model the probability of the fruit being a lemon given its radius?
No! These logits can be any real number, but we want to model the probability of the fruit being a lemon given its radius, which is a number between 0 and 1.
We can use the sigmoid function to map the logits to a number between 0 and 1.
\[ \sigma(x) = \frac{1}{1 + e^{-x}} \]
| radius | logits | probabilities | |
|---|---|---|---|
| 0 | 2.646808 | 1.176169 | 0.764258 |
| 1 | 2.745012 | 1.294014 | 0.784826 |
| 2 | 1.148591 | -0.621690 | 0.349397 |
| 3 | 2.877917 | 1.453500 | 0.810537 |
| 4 | 1.171345 | -0.594386 | 0.355629 |
| radius | logits | probabilities | |
|---|---|---|---|
| 29 | 0.018481 | -1.977822 | 0.121551 |
| 50 | 0.015137 | -1.981835 | 0.121123 |
| 74 | 0.012186 | -1.985377 | 0.120747 |
| 99 | 0.048608 | -1.941670 | 0.125464 |
| 108 | 0.187182 | -1.775382 | 0.144874 |
We can observe as per our model, smaller fruits are more likely to be limes (probability of being a lemon is less) and larger fruits are more likely to be lemons (probability of being a lemon is more).
Generate a dataset
Model Training and Results
Our logistic regression successfully learned the underlying relationship! The learned parameters closely match the true parameters used to generate the data.
| radius | logits | probabilities | y_true | |
|---|---|---|---|---|
| 6 | 0.769717 | -1.076339 | 0.254199 | 0.0 |
| 9 | 0.399558 | -1.520531 | 0.179383 | 0.0 |
| 15 | 1.288213 | -0.454144 | 0.388376 | 0.0 |
| 17 | 1.721713 | 0.066056 | 0.516508 | 0.0 |
| 18 | 0.799740 | -1.040312 | 0.261090 | 0.0 |
| 19 | 1.882347 | 0.258817 | 0.564345 | 0.0 |
| 24 | 0.315945 | -1.620866 | 0.165085 | 0.0 |
| 25 | 0.808484 | -1.029819 | 0.263119 | 0.0 |
| 26 | 1.076438 | -0.708274 | 0.329980 | 0.0 |
| 28 | 1.641575 | -0.030110 | 0.492473 | 0.0 |
| radius | logits | probabilities | y_true | |
|---|---|---|---|---|
| 0 | 2.646808 | 1.176169 | 0.764258 | 1.0 |
| 1 | 2.745012 | 1.294014 | 0.784826 | 1.0 |
| 2 | 1.148591 | -0.621690 | 0.349397 | 1.0 |
| 3 | 2.877917 | 1.453500 | 0.810537 | 1.0 |
| 4 | 1.171345 | -0.594386 | 0.355629 | 1.0 |
| 5 | 1.802686 | 0.163223 | 0.540715 | 1.0 |
| 7 | 2.380924 | 0.857109 | 0.702056 | 1.0 |
| 8 | 2.822314 | 1.386777 | 0.800077 | 1.0 |
| 10 | 2.803794 | 1.364553 | 0.796499 | 1.0 |
| 11 | 1.780739 | 0.136887 | 0.534168 | 1.0 |
We can notice that even though the probability of the event is very low, it still happens. This is the nature of the Bernoulli distribution.
| radius | logits | probabilities | y_true | |
|---|---|---|---|---|
| 0 | 2.646808 | 1.176169 | 0.764258 | 0.0 |
| 3 | 2.877917 | 1.453500 | 0.810537 | 0.0 |
| 4 | 1.171345 | -0.594386 | 0.355629 | 0.0 |
| 5 | 1.802686 | 0.163223 | 0.540715 | 0.0 |
| 7 | 2.380924 | 0.857109 | 0.702056 | 0.0 |
| 9 | 0.399558 | -1.520531 | 0.179383 | 0.0 |
| 10 | 2.803794 | 1.364553 | 0.796499 | 0.0 |
| 14 | 2.223282 | 0.667939 | 0.661041 | 0.0 |
| 15 | 1.288213 | -0.454144 | 0.388376 | 0.0 |
| 18 | 0.799740 | -1.040312 | 0.261090 | 0.0 |
# Plot the data
plt.scatter(radius_array, y_true, alpha=0.1, marker='|', color='k')
plt.xlabel('Radius')
# Use Limes and Lemon markers only on y-axis
plt.yticks([0, 1], ['Limes', 'Lemons'])
plt.ylabel('Fruit')Text(0, 0.5, 'Fruit')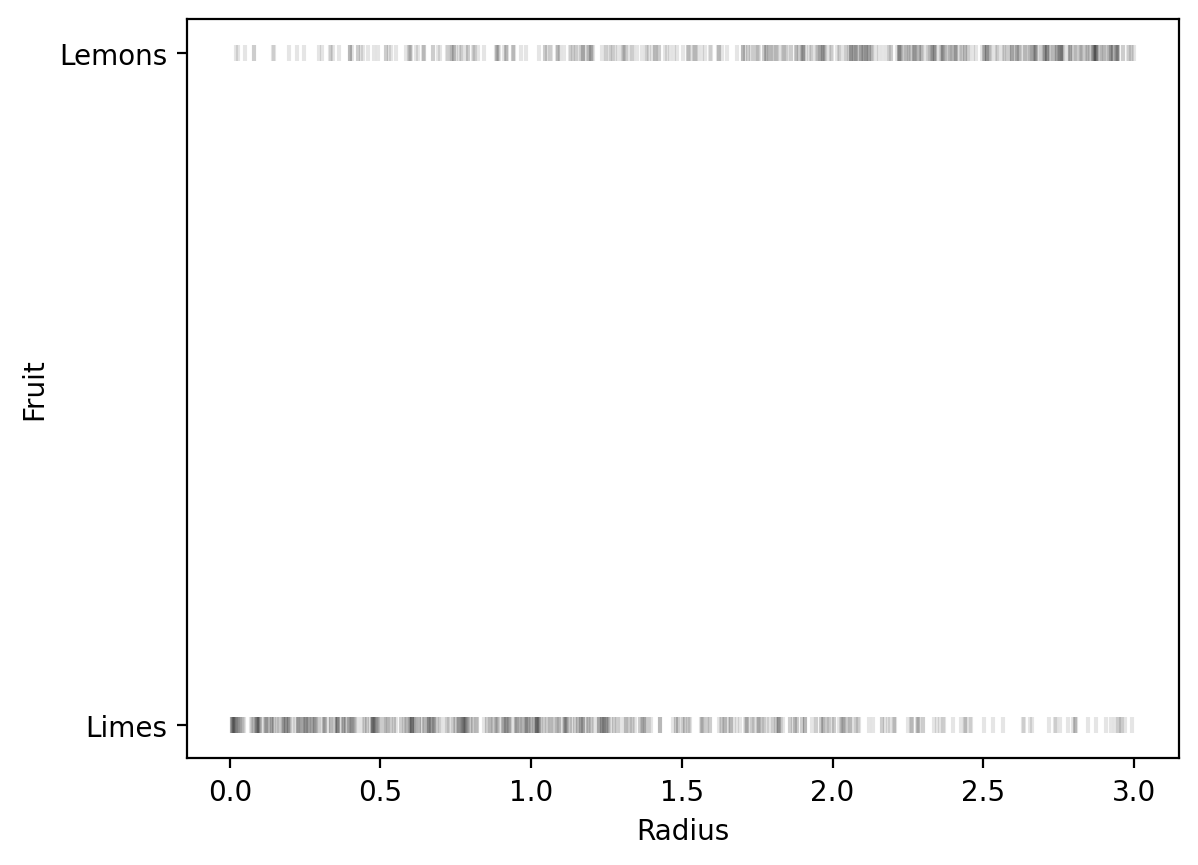
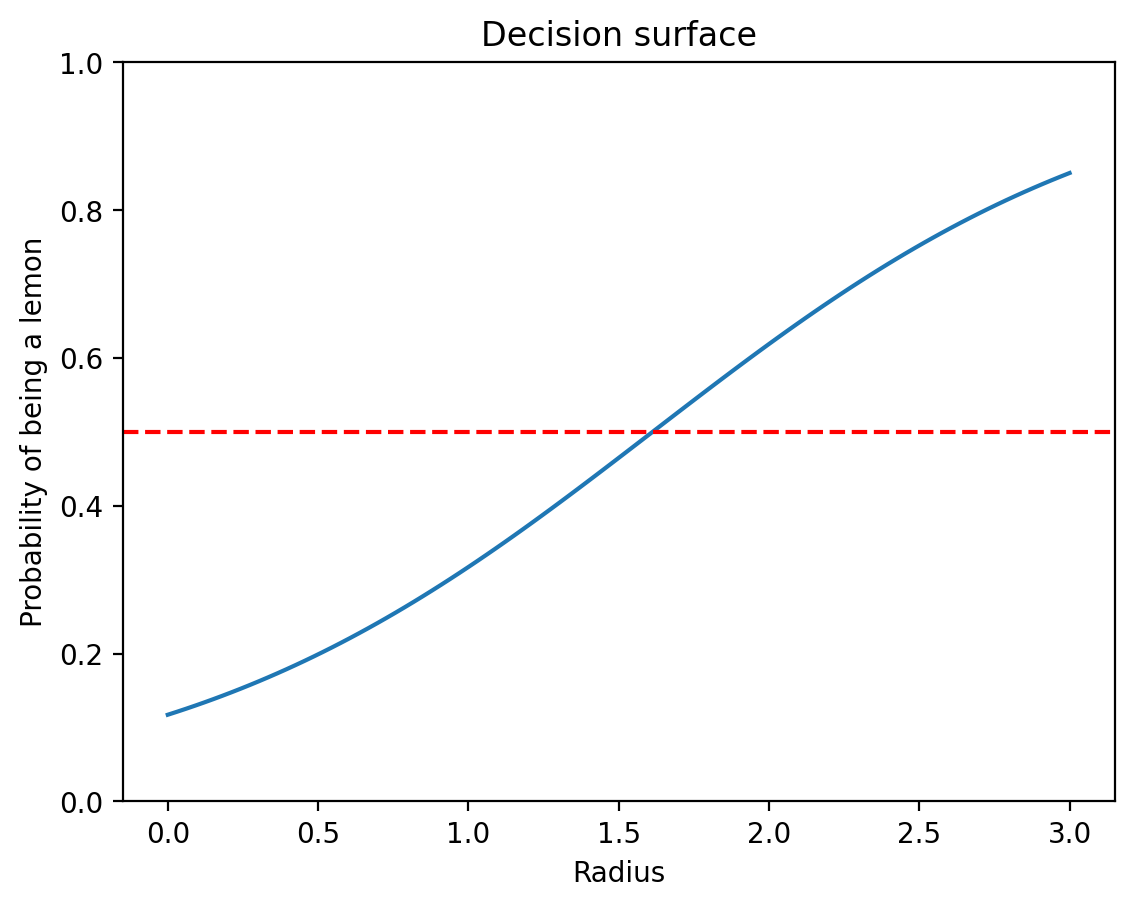
The decision boundary is at radius ≈ 1.67, where P(Lemon) = 0.5.
2. Categorical Distribution
The Categorical distribution generalizes the Bernoulli distribution to more than two outcomes. It’s fundamental for multi-class classification problems.
Mathematical Definition
Let \(X\) be a categorical random variable with \(K\) possible outcomes \(\{1, 2, \ldots, K\}\) and parameter vector \(\boldsymbol{\theta} = (\theta_1, \theta_2, \ldots, \theta_K)\). The PMF is:
\[p_X(x = k) = \theta_k \quad \text{for } k \in \{1, 2, \ldots, K\}\]
Constraints: \(\theta_k \geq 0\) and \(\sum_{k=1}^K \theta_k = 1\)
Applications
- Image classification (cat, dog, bird, …)
- Sentiment analysis (positive, negative, neutral)
- Die rolling (1, 2, 3, 4, 5, 6)
- Market research (product preferences)
# Logistic regression model
class LogisticRegression(nn.Module):
def __init__(self):
super(LogisticRegression, self).__init__()
self.linear = nn.Linear(1, 1)
def forward(self, x):
return self.linear(x)
model = LogisticRegression()
# Training the model
criterion = nn.BCEWithLogitsLoss()
optimizer = torch.optim.Adam(model.parameters(), lr=0.01)
# Convert the data to PyTorch tensors
radius_tensor = radius_array.unsqueeze(1)
y_true_tensor = y_true.unsqueeze(1)
# Training loop
n_epochs = 1000
for epoch in range(n_epochs):
model.train()
optimizer.zero_grad()
# Forward pass
y_pred = model(radius_tensor)
# Compute loss
loss = criterion(y_pred, y_true_tensor)
# Backward pass
loss.backward()
# Update weights
optimizer.step()
if epoch % 100 == 0:
print(f'Epoch: {epoch}, Loss: {loss.item()}')
Epoch: 0, Loss: 0.8052948117256165
Epoch: 100, Loss: 0.6282714605331421
Epoch: 200, Loss: 0.593073308467865
Epoch: 300, Loss: 0.5790674686431885
Epoch: 400, Loss: 0.5744827389717102
Epoch: 500, Loss: 0.5731877088546753
Epoch: 600, Loss: 0.5728737711906433
Epoch: 700, Loss: 0.5728095769882202
Epoch: 800, Loss: 0.5727986693382263
Epoch: 900, Loss: 0.5727971196174622# Learned weights and bias
w_learned = model.linear.weight.item()
b_learned = model.linear.bias.item()
# Compare the true and learned weights and bias
print(f'True weights: {w_true}, Learned weights: {w_learned}')
print(f'True bias: {b_true}, Learned bias: {b_learned}')True weights: 1.2, Learned weights: 1.2514334917068481
True bias: -2.0, Learned bias: -2.0183537006378174# Decision surface
radius_values = torch.linspace(0, 3, 100).unsqueeze(1)
probs = sigmoid(model(radius_values)).detach()
plt.plot(radius_values, probs)
plt.xlabel('Radius')
plt.ylabel('Probability of being a lemon')
plt.title('Decision surface')
plt.ylim(0, 1)
plt.axhline(0.5, color='r', linestyle='--')
Categorical distribution
Say, we have a random variable \(X\) that can take on one of \(K\) possible values, \(1, 2, \ldots, K\). The probability mass function (PMF) of a categorical distribution is given by:
\[ p_X(x) = \begin{cases} \theta_1, & \text{if } x = 1, \\ \theta_2, & \text{if } x = 2, \\ \vdots \\ \theta_K, & \text{if } x = K. \end{cases} \]
where \(\theta_1, \theta_2, \ldots, \theta_K\) are the parameters of the categorical distribution and satisfy the following constraints:
\[ 0 \leq \theta_i \leq 1, \quad \sum_{i=1}^K \theta_i = 1. \]
We write
\[ X \sim \text{Categorical}(\theta_1, \theta_2, \ldots, \theta_K) \]
to denote that \(X\) is drawn from a categorical distribution with parameters \(\theta_1, \theta_2, \ldots, \theta_K\). The categorical distribution is a generalization of the Bernoulli distribution to more than two outcomes.
If we had a fair 6-sided die, the PMF of the die roll would be given by:
\[ p_X(x) = \frac{1}{6}, \quad x \in \{1, 2, 3, 4, 5, 6\}. \]
Imagenet
The ImageNet project is a large visual database designed for use in visual object recognition research.
The empirical frequencies closely match our theoretical probabilities, validating our implementation.
3. Binomial Distribution
The Binomial distribution models the number of successes in a fixed number of independent Bernoulli trials.
Mathematical Definition
Let \(X\) be the number of successes in \(n\) independent Bernoulli trials, each with success probability \(p\). Then \(X \sim \text{Binomial}(n, p)\) with PMF:
\[p_X(x) = \binom{n}{x} p^x (1-p)^{n-x} \quad \text{for } x \in \{0, 1, 2, \ldots, n\}\]
where \(\binom{n}{x} = \frac{n!}{x!(n-x)!}\) is the binomial coefficient.
Properties
- Mean: \(E[X] = np\)
- Variance: \(\text{Var}(X) = np(1-p)\)
- Mode: \(\lfloor (n+1)p \rfloor\)
Real-World Example: Quality Control
A factory produces electronic chips with a 10% defect rate. Quality control randomly selects 10 chips. How many defects should we expect?

theta_vec = torch.tensor([0.1, 0.2, 0.3, 0.4])
#
ser = pd.Series(theta_vec.numpy())
ser.plot(kind='bar', rot=0)
plt.xlabel('Outcome')
plt.ylabel('Probability')Text(0, 0.5, 'Probability')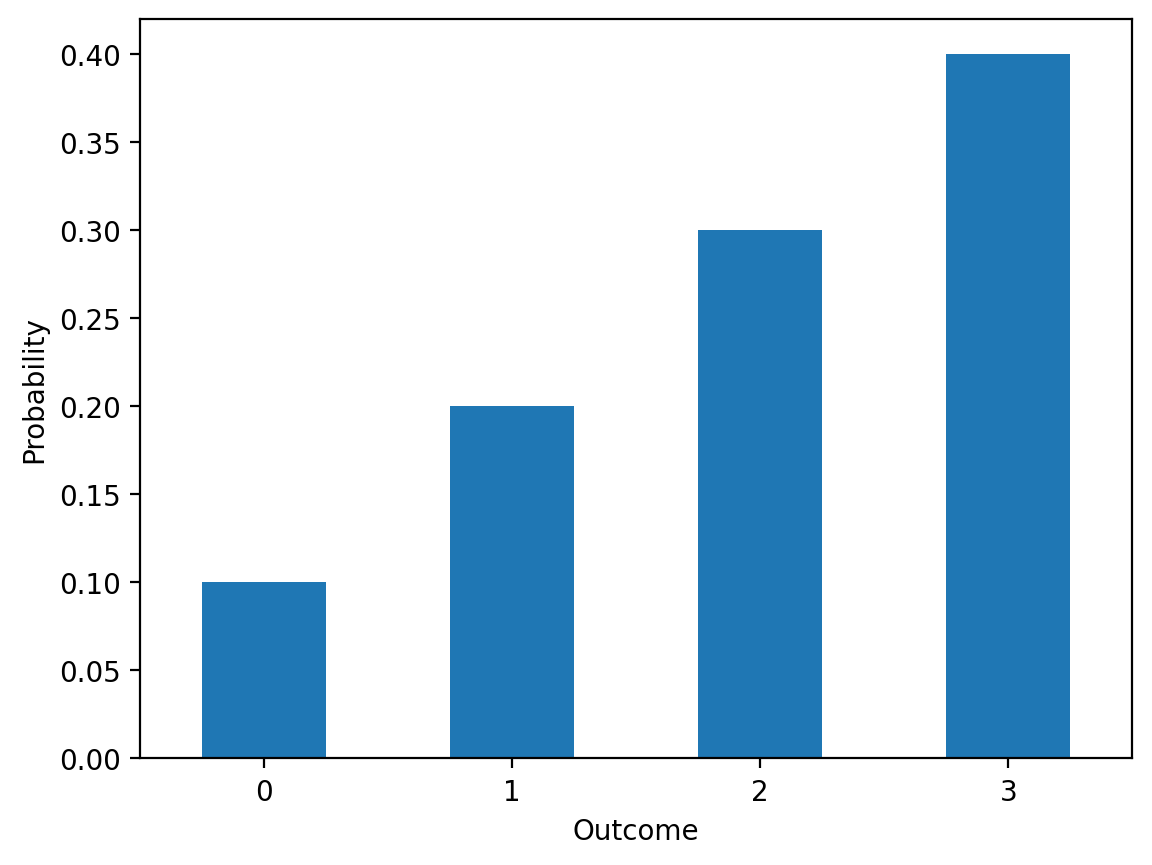
Categorical(probs: torch.Size([4]))Expected value argument (Tensor of shape ()) to be within the support (IntegerInterval(lower_bound=0, upper_bound=3)) of the distribution Categorical(probs: torch.Size([4]), logits: torch.Size([4])), but found invalid values:
4.0The distribution shows that finding 0 or 1 defects is most likely, with very low probability of finding 5+ defects in a sample of 10.
Simulation Validation
4. Poisson Distribution
The Poisson distribution models the number of events occurring in a fixed interval when events happen independently at a constant average rate.
Quality control in factories
A factory produces electronic chips, and each chip has a probability \(p\) of being defective due to manufacturing defects. The quality control team randomly selects 10 chips from a large production batch and checks how many are defective.
How can we model the number of defective chips in the sample?
We can model the number of defective chips in the sample using a binomial distribution.
Let \(X\) be the number of defective chips in the sample. \(X\) can take on values \(0, 1, 2, \ldots, 10\). The probability mass function (PMF) of a binomial distribution is given by:
\[ p_X(x) = \binom{n}{x} p^x(1-p)^{n-x}, \quad x \in \{0, 1, 2, \ldots, 10\}. \]
where \(n\) is the number of chips in the sample, \(0 < p < 1\) is the probability of a chip being defective, and \(\binom{n}{x}\) is the binomial coefficient, which is the number of ways to choose \(x\) defective chips from \(n\) chips.
Mathematical Definition
Let \(X\) be the number of events in a fixed interval. If events occur at rate \(\lambda > 0\), then \(X \sim \text{Poisson}(\lambda)\) with PMF:
\[p_X(x) = \frac{\lambda^x e^{-\lambda}}{x!} \quad \text{for } x \in \{0, 1, 2, \ldots\}\]
Properties
- Mean: \(E[X] = \lambda\)
- Variance: \(\text{Var}(X) = \lambda\)
- Unique property: Mean equals variance!
Applications
- Email arrivals per hour
- Customer arrivals at a store
- Defects per manufactured unit
- Website clicks per minute
- Radioactive decay events
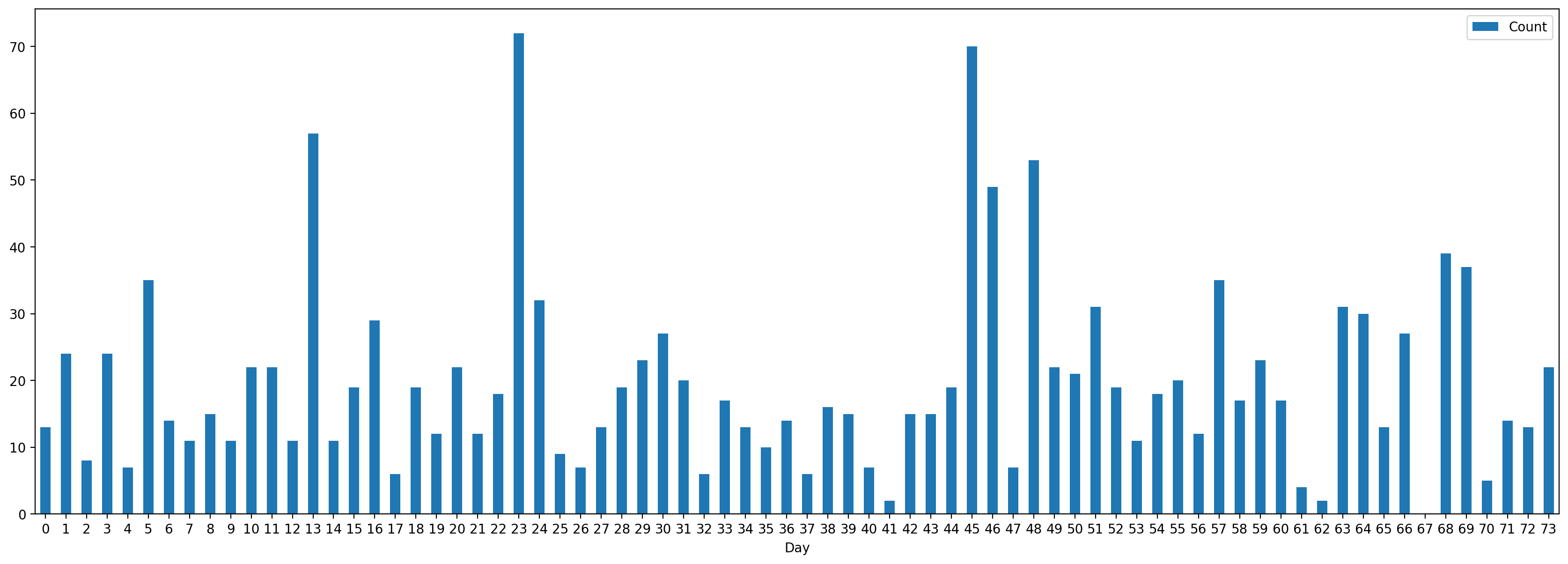
Real-World Example: SMS Message Arrivals
Let’s analyze SMS message data to see if it follows a Poisson distribution:
x = torch.arange(0, n_chips+1)
y = dist.log_prob(x).exp()
df_prob_binomial = pd.DataFrame({
'x': x.numpy(),
'P(X=x)': y.numpy().round(5)
})
df_prob_binomial| x | P(X=x) | |
|---|---|---|
| 0 | 0 | 0.34868 |
| 1 | 1 | 0.38742 |
| 2 | 2 | 0.19371 |
| 3 | 3 | 0.05740 |
| 4 | 4 | 0.01116 |
| 5 | 5 | 0.00149 |
| 6 | 6 | 0.00014 |
| 7 | 7 | 0.00001 |
| 8 | 8 | 0.00000 |
| 9 | 9 | 0.00000 |
| 10 | 10 | 0.00000 |
Text(0, 0.5, 'P(X=x)')
Mathematical Definition
Let \(X\) be the number of trials needed to get the first success, where each trial has success probability \(p\). Then \(X \sim \text{Geometric}(p)\) with PMF:
\[p_X(x) = (1-p)^{x-1} p \quad \text{for } x \in \{1, 2, 3, \ldots\}\]
Note: PyTorch uses a different parameterization where \(X\) counts the number of failures before the first success, so \(X \in \{0, 1, 2, \ldots\}\).
Properties
- Mean: \(E[X] = \frac{1}{p}\) (trials to first success)
- Variance: \(\text{Var}(X) = \frac{1-p}{p^2}\)
- Memoryless property: \(P(X > m+n | X > m) = P(X > n)\)
Applications
- Sales calls until first sale
- Equipment testing until first failure
- Quality control until first defect
- Customer service until problem resolution
Example: Sales Representative
A sales representative has a 30% success rate per call. How many calls until the first sale?
5. Geometric Distribution
The Geometric distribution models the number of trials needed to achieve the first success in a sequence of independent Bernoulli trials.
Number of SMS rceived per day
Hat tip: Bayesian Methods for Hackers book
Remember that PyTorch’s Geometric distribution counts failures before success, so these represent the number of failed calls before the first successful sale.
Summary and Key Insights
In this comprehensive notebook, we’ve explored the fundamental discrete probability distributions that form the backbone of statistical modeling and machine learning:
Distributions Covered
- Bernoulli: Binary outcomes (classification, A/B testing)
- Categorical: Multi-class outcomes (image classification, NLP)
- Binomial: Fixed trials with binary outcomes (quality control, surveys)
- Poisson: Count of rare events (arrivals, defects, clicks)
- Geometric: Trials to first success (sales, reliability testing)
Key Mathematical Insights
- PMF Properties: Non-negativity and normalization are universal
- Parameter Relationships: Mean and variance depend on distribution parameters
- Support: Each distribution has a specific domain of possible values
- Applications: Choice depends on the underlying data-generating process
Practical Implementation Skills
- PyTorch Integration: Using built-in distributions for modeling
- Sampling and Validation: Generating samples and verifying against theory
- Real-World Modeling: Applying distributions to practical problems
- Parameter Estimation: Learning distribution parameters from data
When to Use Each Distribution
| Situation | Distribution | Key Question |
|---|---|---|
| Binary classification | Bernoulli | Success/failure with fixed probability? |
| Multi-class classification | Categorical | Multiple exclusive outcomes? |
| Fixed number of trials | Binomial | How many successes in n trials? |
| Count of rare events | Poisson | How many events in fixed time/space? |
| Time to first success | Geometric | How long until first success? |
Advanced Topics for Further Study
- Maximum Likelihood Estimation for parameter fitting
- Bayesian approaches to parameter uncertainty
- Mixture models combining multiple distributions
- Zero-inflated models for count data with excess zeros
- Truncated distributions for bounded domains
Machine Learning Connections
These distributions are essential for: - Generative models (VAEs, GANs) - Classification (logistic regression, neural networks) - Reinforcement learning (policy gradients) - Bayesian machine learning (priors and posteriors) - Natural language processing (language models)
Understanding discrete distributions provides the foundation for advanced statistical modeling and machine learning applications!
| Count | |
|---|---|
| Day | |
| 0 | 13.0 |
| 1 | 24.0 |
| 2 | 8.0 |
| 3 | 24.0 |
| 4 | 7.0 |
| ... | ... |
| 69 | 37.0 |
| 70 | 5.0 |
| 71 | 14.0 |
| 72 | 13.0 |
| 73 | 22.0 |
74 rows × 1 columns
How can you model the number of SMS messages you receive per day?
We can model the number of SMS messages you receive per day using a Poisson distribution.
Let \(X\) be the number of SMS messages you receive per day. \(X\) can take on values \(0, 1, 2, \ldots\). The probability mass function (PMF) of a Poisson distribution is given by:
\[ p_X(x) = \frac{\lambda^x e^{-\lambda}}{x!}, \quad x \in \{0, 1, 2, \ldots\}. \]
where \(\lambda > 0\) is the average number of SMS messages you receive per day.
x_range = torch.arange(0, 20)
y = dist.log_prob(x_range).exp()
df_prob_poisson = pd.DataFrame({
'x': x_range.numpy(),
'P(X=x)': y.numpy().round(6)
})
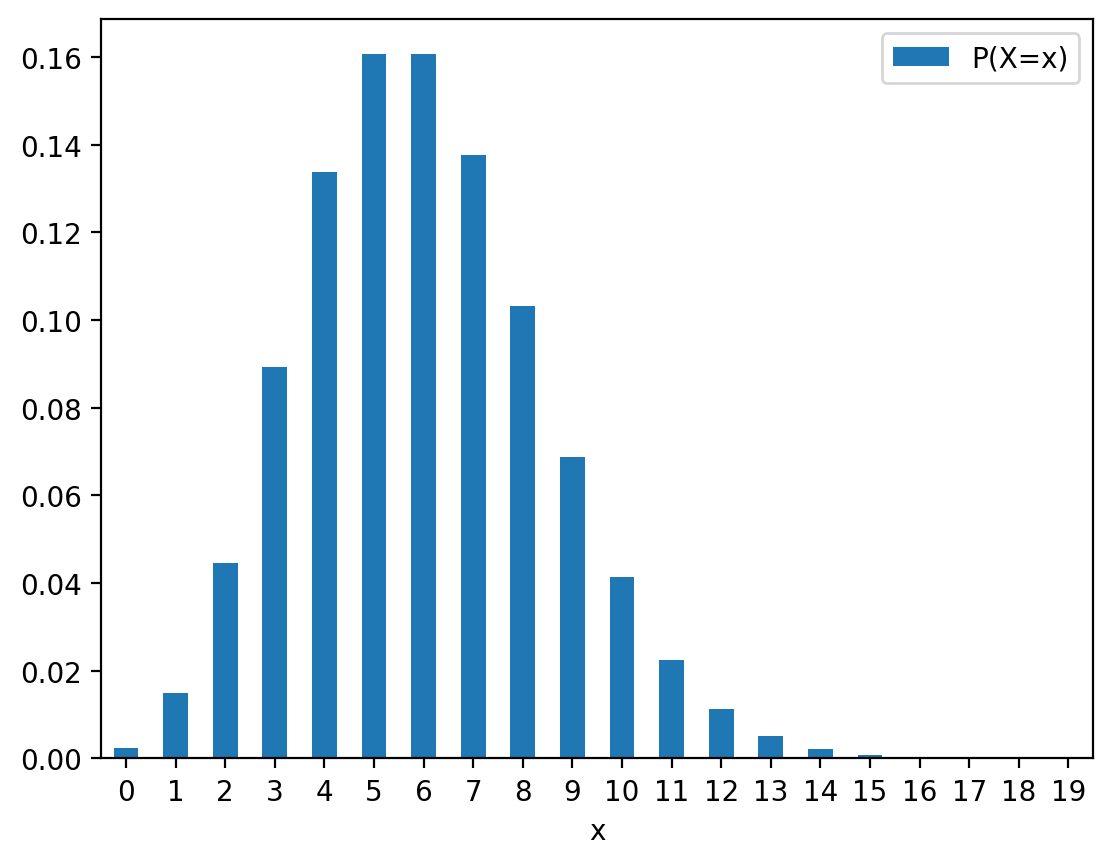
df_prob_poisson| x | P(X=x) | |
|---|---|---|
| 0 | 0 | 0.002479 |
| 1 | 1 | 0.014873 |
| 2 | 2 | 0.044618 |
| 3 | 3 | 0.089235 |
| 4 | 4 | 0.133853 |
| 5 | 5 | 0.160623 |
| 6 | 6 | 0.160623 |
| 7 | 7 | 0.137677 |
| 8 | 8 | 0.103258 |
| 9 | 9 | 0.068839 |
| 10 | 10 | 0.041303 |
| 11 | 11 | 0.022529 |
| 12 | 12 | 0.011264 |
| 13 | 13 | 0.005199 |
| 14 | 14 | 0.002228 |
| 15 | 15 | 0.000891 |
| 16 | 16 | 0.000334 |
| 17 | 17 | 0.000118 |
| 18 | 18 | 0.000039 |
| 19 | 19 | 0.000012 |
Sales Calls Before a Successful Sale
A sales representative makes cold calls to potential customers. Each call has a probability \(p\) of resulting in a successful sale. We want to model the number of calls needed before achieving a successful sale.
We can model the number of calls needed before achieving a successful sale using a geometric distribution.
Let \(X\) be the number of calls needed before achieving a successful sale. \(X\) can take on values \(1, 2, 3, \ldots\). The probability mass function (PMF) of a geometric distribution is given by:
\[ p_X(x) = (1-p)^{x-1}p, \quad x \in \{1, 2, 3, \ldots\}. \]
where \(0 < p < 1\) is the probability of a successful sale on each call.